Dimension conversion and scaling of disordered protein chains†
Abstract
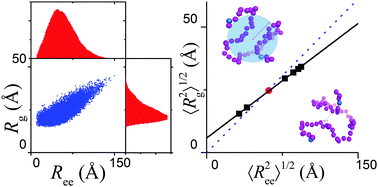
To extract protein dimension and energetics information from single-molecule fluorescence resonance energy transfer spectroscopy (smFRET) data, it is essential to establish the relationship between the distributions of the radius of gyration (Rg) and the end-to-end (donor-to-acceptor) distance (Ree). Here, we performed a coarse-grained molecular dynamics simulation to obtain a conformational ensemble of denatured proteins and intrinsically disordered proteins. For any disordered chain with fixed length, there is an excellent linear correlation between the average values of Rg and Ree under various solvent conditions, but the relationship deviates from the prediction of a Gaussian chain. A modified conversion formula was proposed to analyze smFRET data. The formula reduces the discrepancy between the results obtained from FRET and small-angle X-ray scattering (SAXS). The scaling law in a coil–globule transition process was examined where a significant finite-size effect was revealed, i.e., the scaling exponent may exceed the theoretical critical boundary [1/3, 3/5] and the prefactor changes notably during the transition. The Sanchez chain model was also tested and it was shown that the mean-field approximation works well for expanded chains.

 Please wait while we load your content...
Please wait while we load your content...