Complexation of short ds RNA/DNA oligonucleotides with Gemini micelles: a time resolved SAXS and computational study†
Abstract
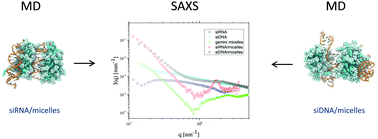
Gene therapy is based on nucleic acid delivery to pathogenic cells in order to modulate their gene expression. The most used non viral vectors are lipid-based nanoaggregates, which are safer than viral carriers and have been shown to assemble easily with both DNA and RNA. However, the transfection efficiency of non viral carriers still needs to be improved before intensive practise in clinical trials can be implemented. For this purpose, the in depth characterization of the complexes formed by nucleic acids and their transporters is of great relevance. In particular, information on the structure and assembly mechanism can be useful to improve our general knowledge of these artificial transfection agents. In this paper, the complexation mechanism of short interfering RNA and DNA molecules (siRNA and siDNA, respectively) with cationic micelles is investigated by combining small angle X-ray scattering experiments and molecular dynamics simulations. Micelles were obtained by Gemini surfactants with different spacer lengths (12-3-12, 12-6-12). The siRNA and siDNA used were double strand molecules characterized by the same length and homologous sequence, in order to perform a close comparison. We showed that complexes appear in solution immediately after mixing and, therefore, the investigation of complex formation requires fast experimental techniques, such as time resolved synchrotron SAXS (Tr-SAXS). The obtained systems had internal arrangement constituted by layers of squeezed micelles alternating the nucleic acids. Both SAXS and MD analyses allowed us to evaluate the mean size of complexes in the range of a few nanometers, with looser and less ordered stacking for the DNA containing aggregates.


 Please wait while we load your content...
Please wait while we load your content...