Characterization of the excited states of DNA building blocks: a coupled cluster computational study†
Abstract
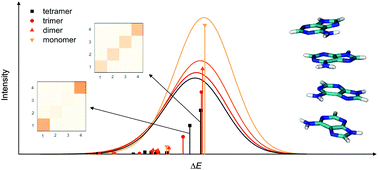
DNA building blocks consisting of up to four nucleobases are investigated using the EOM-CCSD and CC2-LR methods in two B-DNA-like arrangements of a poly-adenine:poly-thymine (poly-A:poly-T) system. Excitation energies and oscillator strengths are presented and the characteristics of the excited states are discussed. Excited states of single-stranded poly-A systems are highly delocalized, especially the spectroscopically bright states, where delocalization over up to four fragments can be observed. In the case of poly-T systems, the states are somewhat less delocalized, extending to maximally about three fragments. A single A:T Watson–Crick pair has highly localized states, while delocalization over base pairs can be observed for some excited states of the (A)2:(T)2 system, but intrastrand delocalization is more pronounced in this case, as well. As for the characteristics of the simulated UV absorption spectra, a significant decrease of intensity can be observed in the case of single strands with increasing chain length; this is due to the stacking interactions and is in accordance with previous results. On the other hand, the breaking of H-bonds between the two strands does not alter the spectral intensity considerably, it only causes a redshift of the absorption band, thus it is unable to explain the experimentally observed DNA hyperchromism on its own, and stacking interactions need to be considered for the description of this effect as well.


 Please wait while we load your content...
Please wait while we load your content...