Atomic scale insights into urea–peptide interactions in solution†
Abstract
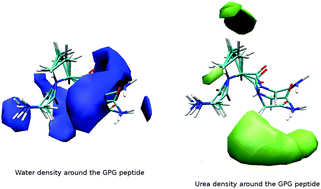
The mechanism by which proteins are denatured by urea is still not well understood, especially on the atomic scale where these interactions occur in vivo. In this study, the structure of the peptide GPG has been investigated in aqueous urea solutions in order to understand the combination of roles that both urea and water play in protein unfolding. Using a combination of neutron diffraction enhanced by isotopic substitution and computer simulations, it was found, in opposition with previous simulations studies, that urea is preferred over water around polar and charged portions of the peptides. Further, it appears that while urea directly replaces water around the nitrogen groups on GPG that urea and water occupy different positions around the peptide bond carbonyl groups. This suggests that urea may in fact weaken the peptide bond, disrupting the peptide backbone, thus ultimately causing denaturation.


 Please wait while we load your content...
Please wait while we load your content...