Analysis of dissolved organic matter fluorescence using self-organizing maps: mini-review and tutorial†
Abstract
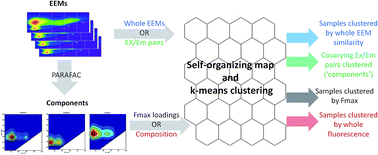
The analysis of dissolved organic matter (DOM) fluorescence has facilitated tracing throughout both natural and engineered aquatic systems, and the connection of DOM quality to its effectiveness as a binding agent, nutrient source, and generator of disinfection byproducts. While methods that deconvolute fluorescence into distinct underlying components have been instrumental in this endeavour, the vast majority of subsequent analyses has been limited to isolated parts/components, whereas DOM is a complex and dynamic system of interacting molecules. Tools for pattern recognition such as Kohonen's self-organizing maps (SOMs) are capable of analyzing DOM fluorescence holistically, either by recomposing the parts isolated via PARAFAC, or by analyzing fluorescence in its entirety; however, the SOM has been underutilized in this endeavour, largely due to the difficulty of implementation and optimization. This report briefly summarizes the prior application of the SOM to DOM fluorescence, followed by a brief introduction to its theory and utility. An instructional tutorial and user-friendly ‘Fluor_SOmap’ Matlab package have been included to assist researchers in implementing SOMs. Four modes of fluorescence data analysis are facilitated by the package, including: whole EEMs, excitation–emission wavelength pairs, component scores from PARAFAC, and overall fluorescence composition. It is hoped that this tutorial and the associated package will help researchers implement and interpret SOMs to extend the scope and utility of fluorescence analysis.

 Please wait while we load your content...
Please wait while we load your content...