Abstract
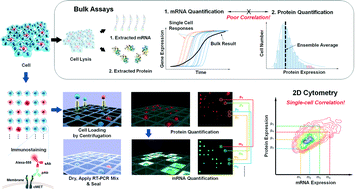
Recently, single-cell molecular analysis has been leveraged to achieve unprecedented levels of biological investigation. However, a lack of simple, high-throughput single-cell methods has hindered in-depth population-wide studies with single-cell resolution. We report a microwell-based cytometric method for simultaneous measurements of gene and protein expression dynamics in thousands of single cells. We quantified the regulatory effects of transcriptional and translational inhibitors on cMET mRNA and cMET protein in cell populations. We studied the dynamic responses of individual cells to drug treatments, by measuring cMET overexpression levels in individual non-small cell lung cancer (NSCLC) cells with induced drug resistance. Across NSCLC cell lines with a given protein expression, distinct patterns of transcript–protein correlation emerged. We believe this platform is applicable for interrogating the dynamics of gene expression, protein expression, and translational kinetics at the single-cell level – a paradigm shift in life science and medicine toward discovering vital cell regulatory mechanisms.

 Please wait while we load your content...
Please wait while we load your content...