The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis†
Abstract
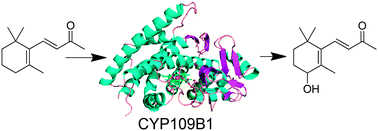
The crystal structure of the versatile CYP109B1 enzyme from Bacillus subtilis has been solved at 1.8 Å resolution. This is the first structure of an enzyme from this CYP family, whose members are prevalent across diverse species of bacteria. In the crystal structure the enzyme has an open conformation with an access channel leading from the heme to the surface. The substrate-free structure reveals the location of the key residues in the active site that are responsible for binding the substrate in the correct orientation for regioselective oxidation. Importantly, there are significant differences among these residues in members of the CYP109 and closely related CYP106 families and these likely account for the variations in substrate binding and oxidation profiles observed with these enzymes. A whole-cell oxidation biosystem was developed, which contains CYP109B1 and a phthalate family oxygenase reductase (PFOR), from Pseudomonas putida KT24440, as the electron transfer partner. This electron transfer system is able to support CYP109B1 activity resulting in the regioselective hydroxylation of both α- and β-ionone in vivo and in vitro. The PFOR is therefore a versatile electron transfer partner that is able to support the activity of CYP enzymes from other bacterium. The crystal structure of CYP109B1 has a positively charged proximal face and this explains why it can interact with PFOR and adrenodoxin which are predominantly negatively charged around their [2Fe–2S] clusters.
- This article is part of the themed collection: Chemical Biology in Molecular BioSystems

 Please wait while we load your content...
Please wait while we load your content...