Multi-omics based changes in response to cadmium toxicity in Bacillus licheniformis A
Abstract
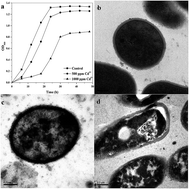
Cadmium (Cd), a widespread substance with high toxicity and persistence, is known to cause a broad range of adverse effects in all living organisms. However, many microorganisms have special detoxification mechanisms for Cd. In this study, we investigated the response to Cd in Bacillus licheniformis A at the omics level. Proteomic biomarkers were identified, including peroxiredoxin, nucleoside diphosphate kinase, trigger factor, and elongation factor Tu. These proteins suggested that Cd could induce oxidative stress, disturbance in energy metabolism, and protein synthesis in B. licheniformis A. Furthermore, metabolites (glutamate, glycine, and S-adenosylhomocysteine) involved in the glutathione synthesis pathway were also affected. In addition, some metabolic biomarkers were validated by relating to respective proteins, including 50S ribosomal protein L7/L12, trigger factor, peroxiredoxin and superoxide dismutase in B. licheniformis A. This study provides new insights into the utilization of an omics approach in bacteria to assess the effects of Cd pollution in the environment.

 Please wait while we load your content...
Please wait while we load your content...