Mathematical modeling deciphers the benefits of alternatively-designed conserved activatory and inhibitory gene circuits†
Abstract
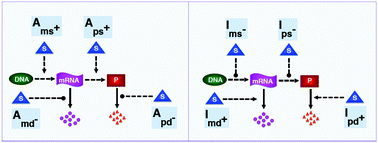
Cells employ a variety of mechanisms as a response to external signals to maintain cellular homeostasis. In this study, we examine four activatory and four inhibitory protein synthesis mechanisms at both population and single cell level that can be triggered by a transient external signal. Activation mechanisms result from the assumption that cells can employ four different modes to temporarily increase the levels of a protein: decreased mRNA degradation, increased mRNA synthesis, decreased protein degradation and increased protein synthesis. For the inhibition mechanisms it is assumed that a cell can reduce a protein's level through four ways: increased mRNA degradation, reduced mRNA synthesis, increased protein degradation and reduced protein synthesis. Deterministic and stochastic models were developed to analyze the dynamic responses of these eight mechanisms to a transient signal. Three different response metrics were used to measure different aspects of the response. These metrics are (i) mid-protein abundance (mP), (ii) time required for the protein to reach the mid-protein level (mT), and (iii) duration of response (D), which is defined as the total time for which the protein (P) abundance are above or below of mid-protein level. Our simulations show that of the activation mechanisms, the signal-dependent increase in mRNA synthesis and protein synthesis are more effective and faster, than the signal dependent decrease in mRNA and protein degradation. On the other hand, the mechanism involving signal dependent increase in protein synthesis is noisier than the signal dependent increase in mRNA synthesis in regard to all metrics used. Of the four inhibition mechanisms, the signal-dependent increase in the protein degradation is the most effective and fastest of the four inhibition mechanisms. It is also noisiest of the four inhibition mechanisms before the protein levels reach a steady state around 100 minutes.

 Please wait while we load your content...
Please wait while we load your content...