Two polypyridyl copper(ii) complexes: synthesis, crystal structure and interaction with DNA and serum protein in vitro†
Abstract
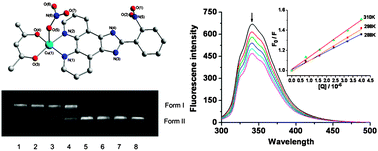
Two polypyridyl copper(II) complexes, [Cu(acac)(p-CPIP)(CH3OH)](NO3) (1) and [Cu(acac)(o-NPIP)(NO3)] (2) (acac = acetylacetonate, p-CPIP = 2-(4-chlorophenyl)imidazo[4,5-f]1,10-phenanthroline, o-NPIP = 2-(2-nitrophenyl)imidazo[4,5-f]1,10-phenanthroline), have been synthesized and characterized by single-crystal analysis, IR and electronic spectroscopy. X-ray analyses reveal that both 1 and 2 possess a triclinic crystal system with Pmn1 and mononuclear five-coordinated square pyramidal geometry, which were further linked to a one-dimensional structure through π–π stacking and intermolecular hydrogen bonds. The interaction of the Cu(II) complex with calf thymus DNA (CT-DNA) was investigated by UV-visible and fluorescence emission spectrometry, as well as agarose gel electrophoresis. The apparent binding constant (Kapp) values of 2.47 × 104 M−1 for 1 and 3.04 × 104 M−1 for 2 suggest moderate intercalative binding mode between the complexes and DNA. Both complexes displayed efficient oxidative cleavage of supercoiled DNA in the presence of external agents. In addition, fluorescence spectrometry of bovine serum albumin (BSA) with the complexes showed that the quenching mechanism of 1 might be a dynamic procedure, while 2 showed a combined dynamic and static quenching mechanism. For both 1 and 2, the number of binding sites was about one for BSA. Moreover, synchronous fluorescence spectral experiments revealed that 1 and 2 affect the microenvironment of tryptophan residues of BSA.

 Please wait while we load your content...
Please wait while we load your content...