Repair of methyl lesions in RNA by oxidative demethylation
Abstract
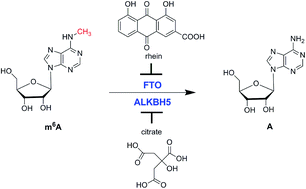
Nucleic acid methylation is one of the most important epigenetic modifications that have been studied intensively for the past several decades. Many diseases, such as cancer, neurological diseases and developmental defects, are closely associated with alkylation damage of nucleic acids. Although the majority of research on nucleic acid modification involves DNA methylation, RNA is also extensively modified. RNA methylation is a common and naturally occurring modification in both prokaryotic and eukaryotic organisms. The methylation of N6-methyladenine (m6A) is the most abundant modification in RNA and plays a fundamental regulatory role in gene expression. Alkylation damage in RNA can be repaired by RNA demethylases, including Escherichia coli AlkB and human AlkB homologues ALKBH1, ALKBH3, ALKBH5 and FTO. ALKBH5 and FTO are the only two m6A RNA demethylases identified so far. In this review, recent findings on RNA demethylases as well as biological functions and inhibitors will be discussed.

 Please wait while we load your content...
Please wait while we load your content...