Exploring the substrate promiscuity of an antibiotic inactivating enzyme†
Abstract
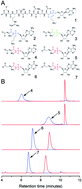
Peptide–nucleotide conjugates have been extensively studied as scaffolds for the development of new antibiotics. However, in vivo, the efficacy of such compounds is limited by various detoxicants, such as aminoacyl-nucleotide hydrolase MccF. MccF cleaves the amide bond between amino acid and phosphoramine–adenylate of the aspartyl tRNA synthetase inhibitor microcin C7, providing self-immunity to the producing strains. However, MccF orthologs are also found in strains that do not produce microcin C7, suggesting a broader role in detoxification. Here, we demonstrate that MccF has no specificity for the nucleotide moiety of the antibiotic and can accept amino acids linked to any purine nucleobase as substrates. Biochemical characterization of synthetic substrate analogs and the co-crystal structure of these compounds with MccF provide a rationale for understanding this promiscuity. These findings have implications for the design of antibiotics that can avert MccF-mediated inactivation and for understanding the function of homologs that may play roles in the metabolism of other cellular intermediates.

 Please wait while we load your content...
Please wait while we load your content...