KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model†
Abstract
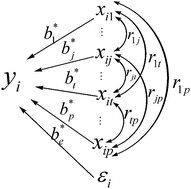
The dynamic impact approach (DIA) represents an alternative to overrepresentation analysis (ORA) for functional analysis of time-course experiments or those involving multiple treatments. The DIA can be used to estimate the biological impact of the differentially expressed genes (DEGs) associated with particular biological functions, for example, as represented by the Kyoto encyclopedia of genes and genomes (KEGG) annotations. However, the DIA does not take into account the correlated dependence structure of the KEGG pathway hierarchy. We have developed herein a path analysis model (KEGG-PATH) to subdivide the total effect of each KEGG pathway into the direct effect and indirect effect by taking into account not only each KEGG pathway itself, but also the correlation with its related pathways. In addition, this work also attempts to preliminarily estimate the impact direction of each KEGG pathway by a gradient analysis method from principal component analysis (PCA). As a result, the advantage of the KEGG-PATH model is demonstrated through the functional analysis of the bovine mammary transcriptome during lactation.

 Please wait while we load your content...
Please wait while we load your content...