Analysis of the mitochondrial proteome of cybrid cells harbouring a truncative mitochondrial DNA mutation in respiratory complex I
Abstract
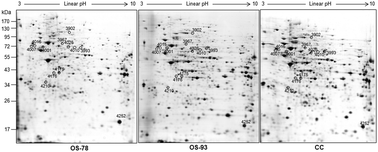
Transmitochondrial cytoplasmic hybrids (cybrids) are well established model systems to reveal the effects of mitochondrial DNA (mtDNA) mutations on cell metabolism excluding the interferences of a different nuclear background. The m.3571insC mutation in the MTND1 gene of respiratory complex I (CI) is commonly detected in oncocytic tumors, in which it causes a severe CI dysfunction leading to an energetic impairment when present above 83% mutant load. To assess whether the energetic deficit may alter the mitochondrial proteome, OS-78 and OS-93 cybrid cell lines bearing two different degrees of the m.3571insC mutation (78% and 92.8%, respectively) and control cybrids bearing wild-type mtDNA (CC) were analyzed. Two-dimensional electrophoresis and mass spectrometry revealed significant alterations only in cybrids above the threshold (OS-93). All differentially expressed proteins are decreased. In particular, the levels of the pyruvate dehydrogenase E1 chain B subunit (E1β), of lipoamide dehydrogenase (E3), the enzyme component of pyruvate and 2-oxoglutarate dehydrogenase complexes, and of lactate dehydrogenase B (LDHB) were reduced. Moreover, a significant decrease of the pyruvate dehydrogenase complex activity was found when OS-93 cybrid cells were grown in galactose medium, a metabolic condition that forces cells to use respiration. These results demonstrate that the energetic impairment caused by the almost homoplasmic m.3571insC mutation perturbs cellular metabolism leading to a decreased steady state level of components of very important mitochondrial NAD-dependent dehydrogenases.
- This article is part of the themed collection: Italian Proteomics Association, 2014

 Please wait while we load your content...
Please wait while we load your content...