Essential O2-responsive genes of Pseudomonas aeruginosa and their network revealed by integrating dynamic data from inverted conditions†
Abstract
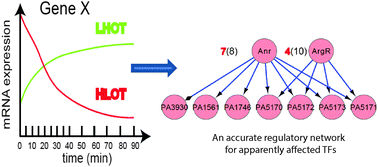
Identification of the gene network through which Pseudomonas aeruginosa PAO1 (PA) adapts to altered oxygen-availability environments is essential for a better understanding of stress responses and pathogenicity of PA. We performed high-time-resolution (HTR) transcriptome analyses of PA in a continuous cultivation system during the transition from high oxygen tension to low oxygen tension (HLOT) and the reversed transition from low to high oxygen tension (LHOT). From those genes responsive to both transient conditions, we identified 85 essential oxygen-availability responsive genes (EORGs), including the expected ones (arcDABC) encoding enzymes for arginine fermentation. We then constructed the regulatory network for the EORGs of PA by integrating information from binding motif searching, literature and HTR data. Notably, our results show that only the sub-networks controlled by the well-known oxygen-responsive transcription factors show a very high consistency between the inferred network and literature knowledge, e.g. 87.5% and 83.3% of the obtained sub-network controlled by the anaerobic regulator (ANR) and a quorum sensing regulator RhIR, respectively. These results not only reveal stringent EORGs of PA and their transcription regulatory network, but also highlight that achieving a high accuracy of the inferred regulatory network might be feasible only for the apparently affected regulators under the given conditions but not for all the expressed regulators on a genome scale.

 Please wait while we load your content...
Please wait while we load your content...