Shedding light on protein folding landscapes by single-molecule fluorescence
Abstract
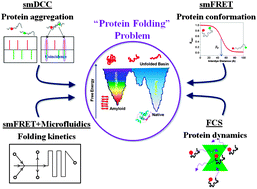
Single-molecule (SM) fluorescence methods have been increasingly instrumental in our current understanding of a number of key aspects of protein folding and aggregation landscapes over the past decade. With the advantage of a model free approach and the power of probing multiple subpopulations and stochastic dynamics directly in a heterogeneous structural ensemble, SM methods have emerged as a principle technique for studying complex systems such as intrinsically disordered proteins (IDPs), globular proteins in the unfolded basin and during folding, and early steps of protein aggregation in amyloidogenesis. This review highlights the application of these methods in investigating the free energy landscapes, folding properties and dynamics of individual protein molecules and their complexes, with an emphasis on inherently flexible systems such as IDPs.
- This article is part of the themed collection: Single-molecule optical spectroscopy

 Please wait while we load your content...
Please wait while we load your content...