Enzymatic combinatorial nucleoside deletion scanning mutagenesis of functional RNA†
Abstract
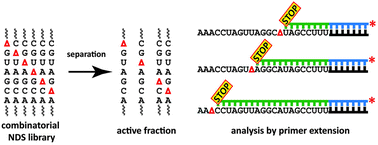
We describe a general and simple method to identify catalytically and structurally important nucleotides in functional RNAs. Our approach is based on statistical replacement of each nucleoside with a non-nucleosidic spacer (C3 linker, Δ), followed by separation of active library variants and readout of interference effects by analysis of enzymatic primer extension reactions.

 Please wait while we load your content...
Please wait while we load your content...