An optimized method for NMR-based plant seed metabolomic analysis with maximized polar metabolite extraction efficiency, signal-to-noise ratio, and chemical shift consistency†
Abstract
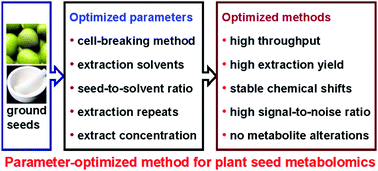
Plant metabolomic analysis has become an essential part of functional genomics and systems biology and requires effective extraction of both primary and secondary metabolites from plant cells. To establish an optimized extraction method for the NMR-based analysis, we used the seeds of mungbean (Vigna radiata cv. Elü no. 1) as a model and systematically investigated the dependence of the metabolite composition in plant extracts on various extraction parameters including cell-breaking methods, extraction solvents, number of extraction repeats, tissue-to-solvent ratio, and extract-to-buffer ratio (for final NMR analysis). We also compared two NMR approaches for quantitative metabolomic analysis from completely relaxed spectra directly and from partially relaxed spectra calculated with T1. By maximizing the extraction efficiency and signal-to-noise ratio but minimizing inter-sample chemical-shift variations and metabolite degradations, we established a parameter-optimized protocol for NMR-based plant seed metabolomic analysis. We concluded that aqueous methanol was the best extraction solvent with an optimal tissue-to-solvent ratio of about 1 : 10–1 : 15 (mg per μL). The combination of tissuelyser homogenization with ultrasonication was the choice of cell-breaking method with three repeated extractions being necessary. For NMR analysis, the optimal extract-to-solvent was around 5–8 mg mL−1 and completely relaxed spectra were ideal for intrinsically quantitative metabolomic analysis although partially relaxed spectra were employable for comparative metabolomics. This optimized method will offer ensured data quality for high-throughput and reliable plant metabolomics studies.

 Please wait while we load your content...
Please wait while we load your content...