Iron-responsive bacterial small RNAs: variations on a theme
Abstract
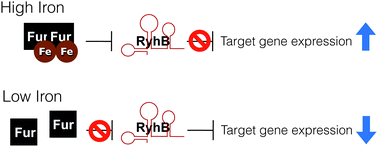
For most living organisms, iron is both essential and potentially toxic, making the precise maintenance of iron homeostasis necessary for survival. To manage this paradox, bacteria regulate the acquisition, utilization, and storage of iron in response to its availability. The iron-dependent ferric uptake repressor (Fur) often mediates this iron-responsive regulation by both direct and indirect mechanisms. In 2002, Masse and Gottesman identified a novel target of Fur-mediated regulation in Escherichia coli: a gene encoding a small regulatory RNA (sRNA) termed RyhB. Under conditions of iron-limitation, RyhB is produced and functions to regulate the expression of several target genes encoding iron-utilizing enzymes, iron acquisition systems, and iron storage factors. This pivotal finding provided the missing link between environmental iron-limitation and previously observed decreases in certain iron-dependent metabolic pathways, a phenomenon now referred to as an “iron-sparing” response. The discovery of RyhB opened the door to the rapidly expanding field of bacterial iron-regulated sRNAs, which continue to be identified and described in numerous bacterial species. Most striking are findings that the impact of iron-responsive sRNA regulation often extends beyond iron homeostasis, particularly with regard to production of virulence-associated factors by pathogenic bacteria. This review discusses trends in the collective body of work on iron-regulated sRNAs, highlighting both the regulatory mechanisms they utilize to control target gene expression and the impact of this regulation on basic processes controlling bacterial physiology and virulence.
- This article is part of the themed collection: Microbial Metallomics

 Please wait while we load your content...
Please wait while we load your content...