RNA-seq data analysis at the gene and CDS levels provides a comprehensive view of transcriptome responses induced by 4-hydroxynonenal†
Abstract
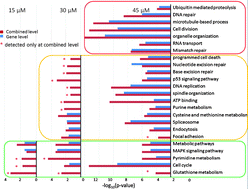
Reactive electrophiles produced during oxidative stress, such as 4-hydroxynonenal (HNE), are increasingly recognized as contributing factors in a variety of degenerative and inflammatory diseases. Here we used the RNA-seq technology to characterize transcriptome responses in RKO cells induced by HNE at subcytotoxic and cytotoxic doses. RNA-seq analysis rediscovered most of the differentially expressed genes reported by microarray studies and also identified novel gene responses. Interestingly, differential expression detection at the coding DNA sequence (CDS) level helped to further improve the consistency between the two technologies, suggesting the utility and importance of the CDS level analysis. RNA-seq data analysis combining gene and CDS levels yielded an informative and comprehensive picture of gradually evolving response networks with increasing HNE doses, from cell protection against oxidative injury at low dose, initiation of cell apoptosis and DNA damage at intermediate dose to significant deregulation of cellular functions at high dose. These evolving dose-dependent pathway changes, which cannot be observed by the gene level analysis alone, clearly reveal the HNE cytotoxic effect and are supported by IC50 experiments. Additionally, differential expression at the CDS level provides new insights into isoform regulation mechanisms. Taken together, our data demonstrate the power of RNA-seq to identify subtle transcriptome changes and to characterize effects induced by HNE through the generation of high-resolution data coupled with differential analysis at both gene and CDS levels.

 Please wait while we load your content...
Please wait while we load your content...