JAK/STAT signalling – an executable model assembled from molecule-centred modules demonstrating a module-oriented database concept for systems and synthetic biology†
Abstract
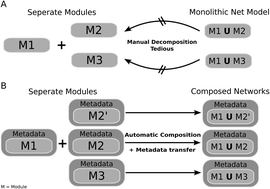
Mathematical models of molecular networks regulating biological processes in cells or organisms are most frequently designed as sets of ordinary differential equations. Various modularisation methods have been applied to reduce the complexity of models, to analyse their structural properties, to separate biological processes, or to reuse model parts. Taking the JAK/STAT signalling pathway with the extensive combinatorial cross-talk of its components as a case study, we make a natural approach to modularisation by creating one module for each biomolecule. Each module consists of a Petri net and associated metadata and is organised in a database publically accessible through a web interface (http://www.biomodelkit.org). The Petri net describes the reaction mechanism of a given biomolecule and its functional interactions with other components including relevant conformational states. The database is designed to support the curation, documentation, version control, and update of individual modules, and to assist the user in automatically composing complex models from modules. Biomolecule centred modules, associated metadata, and database support together allow the automatic creation of models by considering differential gene expression in given cell types or under certain physiological conditions or states of disease. Modularity also facilitates exploring the consequences of alternative molecular mechanisms by comparative simulation of automatically created models even for users without mathematical skills. Models may be selectively executed as an ODE system, stochastic, or qualitative models or hybrid and exported in the SBML format. The fully automated generation of models of redesigned networks by metadata-guided modification of modules representing biomolecules with mutated function or specificity is proposed.

 Please wait while we load your content...
Please wait while we load your content...