Prediction of adverse drug reactions by a network based external link prediction method†
Abstract
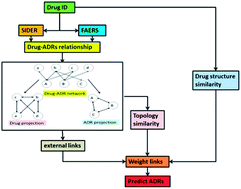
Detecting adverse drug reaction (ADR) is a big challenge to drug development and post-marketing applications. Owing to the low costs and high performance, computational methods are used to predict unknown adverse reactions of drugs. In the present study, a network based method is developed, in which a bipartite network is introduced to represent associations between ADRs and drugs. The potential ADRs of a drug could be simply inferred by its neighbourhood in the bipartite network. Our method was applied on three datasets compiled from FAERS, SIDER and intersection of these two databases (gold standard data). Encouraging results were achieved, area under curve (AUC) values were 0.93, 0.94 and 0.83, respectively. To further evaluate the performance of our method, comparisons were made with internal link prediction method and logistic regression method on the gold standard data. Our method achieved an AUC value of 0.83, while the AUC values were 0.75 for both internal link prediction method and logistic regression method. The results show that it is feasible to predict unknown drug–ADR associations using only topology features of the drug–ADR network.

 Please wait while we load your content...
Please wait while we load your content...