Mutational analysis of phenolic acid decarboxylase from Bacillus subtilis (BsPAD), which converts bio-derived phenolic acids to styrene derivatives†
Abstract
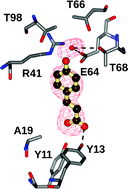
Phenolic acid decarboxylase from Bacillus subtilis (BsPAD) catalyses the decarboxylation of phenolic acids such as coumaric acid to give vinyl phenols, which are of interest as possible polymer precursors and flavour/fragrance compounds. The structure of the Tyr19Ala mutant of BsPAD has been solved in complex with coumaric acid. In the active site, the substrate carboxylate is bound by Tyr11 and Tyr13, and the phenolic hydroxyl by the NE atom of Arg41. A comparison of the mutant complex with the wild-type apoenzyme reveals that the β1–β2 loop, running from Tyr11 to Ala19, closes over the active site in the presence of substrate, shielding it from bulk solvent. The complex structure, in conjunction with an activity study of point mutants of BsPAD, provides support for a mechanism for PADs, proposed by Mancheño and co-workers for the homologue from Lactobacillus plantarum [Proteins, 2010, 78, 1662–1676]. In this mechanism, a quinone methide intermediate results from deprotonation of the phenolic hydroxyl of the substrate by Glu64, assisted by Arg41. Decarboxylation of the substrate is effected through binding of the carboxylate by Tyr11 and Tyr13, the latter being brought into contact with the substrate as a result of the movement of the β1–β2 loop on substrate binding.
- This article is part of the themed collection: Biocatalysis

 Please wait while we load your content...
Please wait while we load your content...