Automated analysis of single stem cells in microfluidic traps†
Abstract
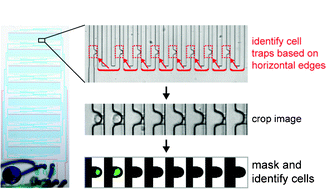
We report a reliable strategy to perform automated image cytometry of single (non-adherent) stem cells captured in microfluidic traps. The method rapidly segments images of an entire microfluidic chip based on the detection of horizontal edges of microfluidic channels, from where the position of the trapped cells can be derived and the trapped cells identified with very high precision (>97%). We used this method to successfully quantify the efficiency and spatial distribution of single-cell loading of a microfluidic chip comprised of 2048 single-cell traps. Furthermore, cytometric analysis of trapped primary hematopoietic stem cells (HSC) faithfully recapitulated the distribution of cells in the G1 and S/G2-M phase of the cell cycle that was measured by

 Please wait while we load your content...
Please wait while we load your content...