Paths of long-range communication in the E2 enzymes of family 3: a molecular dynamics investigation†
Abstract
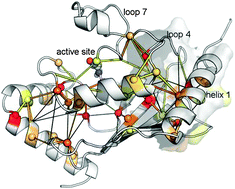
Molecular dynamics (MD) simulations have the ability to help reveal the relationship between protein structure, dynamics and function. Here, we describe MD simulations of the representative members of family 3 of E2 enzymes that we performed and analyzed with the aim of providing a quantitative description of the functional dynamics in this biologically important set of
- This article is part of the themed collection: Theoretical chemical physics of biological systems

 Please wait while we load your content...
Please wait while we load your content...