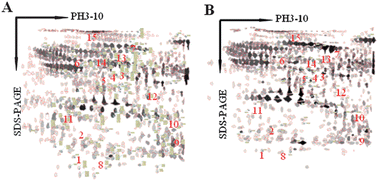
Recent advances in proteomic technologies have enabled us to create detailed protein–protein interaction maps in diseases. As the size of the interaction dataset increases, powerful computational methods are required in order to effectively interpret network models from large scale interactome data. In this study, we carried out comparative proteomics to construct and identify the proteins networks associated with hepatic injury (HI) which are largely unknown, as a case study. All proteins expressed were separated and identified by two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption/ionization time-of-flight-time-of-flight mass spectrometry (MALDI-TOF/TOF MS). Protein-interacting networks and pathways were mapped using STRING analysis program. We have performed for the first time a comprehensive profiling of changes in protein expression of HI rats, to uncover the networks altered by treated with CCl4. Identification of fifteen spots (seven over-expressed and eight under-expressed) were established by MALDI-TOF/TOF MS. These proteins were subjected to functional pathway analysis using STRING software for better understanding of the biological context of the identified proteins. It suggested that modulation of multiple vital physiological pathways including DNA repair process, cell apoptosis, oxidation reduction, signal transduction, metabolic process, intracellular signaling cascade, regulation of biological processes, cell communication, regulation of cellular process, and molecular transport. In summary, the present study provides the first protein-interacting network maps and novel insights into the biological responses and potential pathways of HI. The generation of protein interaction networks clearly enhances the interpretation of proteomic data, particularly in respect of understanding molecular mechanisms of panel protein biomarkers.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...