A parallel algorithm for reverse engineering of biological networks
Abstract
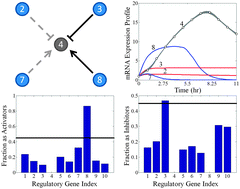
Dynamic biological systems, such as gene regulatory networks (GRNs) and
* Corresponding authors
a
Medical College of Wisconsin, 8701 Watertown Plank Rd., Milwaukee, USA
E-mail:
beardda@gmail.com
Fax: +1 414 955 6568
Tel: +1 456 5752
Dynamic biological systems, such as gene regulatory networks (GRNs) and

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
