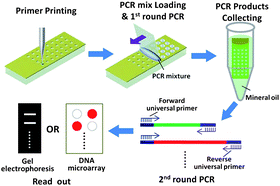
Both basic research and clinical medicine have urgent demands for highly efficient strategies to simultaneously identify many different DNA sequences within a single tube. Effective and simultaneous amplification of multiple target sequences is a prerequisite for any successful multiple nucleic acid detection method. Multiplex PCR is one of the best choices for this purpose. However, due to the intrinsic interference and competition among primer pairs in the same tube, multiple rounds of highly empirical optimization procedures are usually required to establish a successful multiplex PCR reaction. To address this challenge, we report here a universal multiplex PCR strategy that is capable of over 100-plex amplification using a specially designed microarray in which hydrophilic microwells are patterned on a hydrophobic chip. On such an array, primer pairs tagged with a universal sequence are physically separated in individual hydrophilic microwells on an otherwise hydrophobic chip, enabling many unique PCR reactions to be proceeded simultaneously during the first step of the procedure. The PCR products are then isolated and further amplified from the universal sequences, producing a sufficient amount of material for analysis by conventional gel electrophoresis or DNA microarray technology. This strategy is abbreviated as “MPH&HPM” for “![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ultiplex
ultiplex ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) CR on a
CR on a ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ydrophobically and
ydrophobically and ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ydrophilically
ydrophilically ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) atterned
atterned ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) icroarray”. The feasibility of this method is first demonstrated by a multiplex PCR reaction for the simultaneous detection of eleven pneumonia-causing pathogens. Further, we demonstrate the power of this strategy with a highly successful 116-plex PCR reaction that required only little prior optimization. The effectiveness of the MPH&HPM strategy with clinical samples is then illustrated with the detection of deleted exons of the Duchenne Muscular Dystrophy (DMD) gene, the results are in excellent agreement with the clinical records. Because of its generality, simplicity, flexibility, specificity and capacity of more than 100-plex amplification, the MPH&HPM strategy should have broad applications in both laboratory research and clinical applications when multiplex nucleic acid analysis is required.
icroarray”. The feasibility of this method is first demonstrated by a multiplex PCR reaction for the simultaneous detection of eleven pneumonia-causing pathogens. Further, we demonstrate the power of this strategy with a highly successful 116-plex PCR reaction that required only little prior optimization. The effectiveness of the MPH&HPM strategy with clinical samples is then illustrated with the detection of deleted exons of the Duchenne Muscular Dystrophy (DMD) gene, the results are in excellent agreement with the clinical records. Because of its generality, simplicity, flexibility, specificity and capacity of more than 100-plex amplification, the MPH&HPM strategy should have broad applications in both laboratory research and clinical applications when multiplex nucleic acid analysis is required.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) ultiplex
ultiplex ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) CR
CR![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ydrophobically and
ydrophobically and ![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ydrophilically
ydrophilically ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) atterned
atterned ![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) icroarray”. The feasibility of this method is first demonstrated by a multiplex PCR reaction for the simultaneous
icroarray”. The feasibility of this method is first demonstrated by a multiplex PCR reaction for the simultaneous 

 Please wait while we load your content...
Please wait while we load your content...