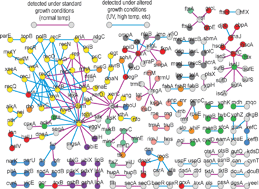
Molecular interactions define the functional organization of the cell. Epistatic (genetic, or gene–gene) interactions, one of the most informative and commonly encountered forms of functional relationships, are increasingly being used to map process architecture in model eukaryotic organisms. In particular, ‘systems-level’ screens in yeast and worm aimed at elucidating genetic interaction networks have led to the generation of models describing the global modular organization of gene products and protein complexes within a cell. However, comparable data for prokaryotic organisms have not been available. Given its ease of growth and genetic manipulation, the Gram-negative bacterium Escherichia coli appears to be an ideal model system for performing comprehensive genome-scale examinations of genetic redundancy in bacteria. In this review, we highlight emerging experimental and computational techniques that have been developed recently to examine functional relationships and redundancy in E. coli at a systems-level, and their potential application to prokaryotes in general. Additionally, we have scanned PubMed abstracts and full-text published articles to manually curate a list of ∼200 previously reported synthetic sick or lethal genetic interactions in E. coli derived from small-scale experimental studies.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...