Functional genomics for plant natural product biosynthesis†
Abstract
Covering: up to February 2009
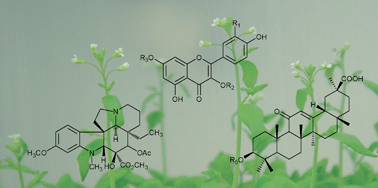
Plants produce an estimated 200,000 secondary metabolites with a vast range of functional and structural diversity. This chemical diversity is due to the multiplicity of the genes in plant genomes. The study of functional genomics based on genomics, transcriptomics and metabolomics is a powerful tool for decoding gene function involved in secondary metabolism in genome-sequenced plants. In particular, substantial progress has been made in Arabidopsis thaliana because of its publicly-available biological materials and genome-related information. A similar strategy can in principle be employed even with exotic plants from which a complete or extensive genomic sequence is not readily available. This review describes the recent developments in functional genomics efforts targeting plant secondary metabolism.
- This article is part of the themed collection: Genomics

 Please wait while we load your content...
Please wait while we load your content...