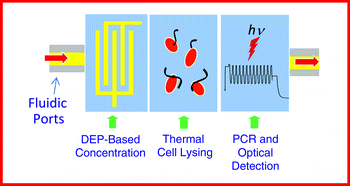
We present a novel, on-chip system for the electrokinetic capture of bacterial cells and their identification using the polymerase chain reaction (PCR). The system comprises a glass–silicon platform with a set of micro-channels, -chambers, and -electrodes. A platinum thin film resistor, placed in the proximity of the chambers, is used for temperature monitoring. The whole chip assembly is mounted on a Printed Circuit Board (PCB) and wire-bonded to it. The PCB has an embedded heater that is utilized for PCR thermal cycle and is controlled by a Lab-View program. Similar to our previous work, one set of electrodes on the chip inside the bigger chamber (0.6 μl volume) is used for diverting bacterial cells from a flowing stream into to a smaller chamber (0.4 nl volume). A second set of interdigitated electrodes (in smaller chamber) is used to actively trap and concentrate the bacterial cells using dielectrophoresis (DEP). In the presence of the DEP force, with the cells still entrapped in the micro-chamber, PCR mix is injected into the chamber. Subsequently, PCR amplification with SYBR Green detection is used for genetic identification of Listeria monocytogenes V7 cells. The increase in fluorescence is recorded with a photomultiplier tube module mounted over an epifluorescence microscope. This integrated micro-system is capable of genetic amplification and identification of as few as 60 cells of L. monocytogenes V7 in less than 90 min, in 600 nl volume collected from a sample of 104 cfu ml−1. Specificity trials using various concentrations of L. monocytogenes V7, Listeria innocua F4248, and Escherichia coli O157:H7 were carried out successfully using two different primer sets specific for a regulatory gene of L. monocytogenes, prfA and 16S rRNA primer specific for the Listeria spp., and no cross-reactivity was observed.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...