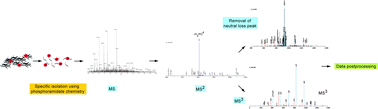
Current methods for phosphoproteome analysis have several limitations. First, most methods for phosphopeptide enrichment lack the specificity to truly purify phosphopeptides. Second, fragmentation spectra of phosphopeptides, in particular those of phosphoserine and phosphothreonine containing peptides, are often dominated by the loss of the phosphate group(s) and therefore lack the information required to identify the peptide sequence and the site of phosphorylation, and third, sequence database search engines and statistical models for data validation are not optimized for the specific fragmentation properties of phosphorylated peptides. Consequently, phosphoproteomic data are characterized by large and unknown rates of false positive and false negative phosphorylation sites. Here we present an integrated chemical, mass spectrometric and computational strategy to improve the efficiency, specificity and confidence in the identification of phosphopeptides and their site(s) of phosphorylation. Phosphopeptides were isolated with high specificity through a simple derivatization procedure based on phosphoramidate chemistry. Identification of phosphopeptides, their site(s) of phosphorylation and the corresponding phosphoproteins was achieved by the optimization of the mass spectrometric data acquisition procedure, the computational tools for database searching and the data post processing. The strategy was applied to the mapping of phosphorylation sites of a purified transcription factor, dFOXO and for the global analysis of protein phosphorylation of Drosophila melanogaster Kc167 cells.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...