 Open Access Article
Open Access ArticleElectrospun protein nanofibers with nanoscale morphological control for dopamine biosensing
Katarzyna Kolodzinska *a,
Sylwia Baluta
*a,
Sylwia Baluta a,
Adrian Cernescub,
Magdalena Wojtasa,
Maciej Lipok
a,
Adrian Cernescub,
Magdalena Wojtasa,
Maciej Lipok a,
Joanna Olesiak-Bańskaa,
Joanna Cabaj
a,
Joanna Olesiak-Bańskaa,
Joanna Cabaj a and
Lech Sznitko
a and
Lech Sznitko *a
*a
aFaculty of Chemistry, Wroclaw University of Science and Technology, Wybrzeze Wyspianskiego 27, 50-370 Wroclaw, Poland. E-mail: katarzyna.kolodzinska@pwr.edu.pl; lech.sznitko@pwr.edu.pl
bNeaspec—Attocube Systems AG, Eglfinger Weg 2, Haar 85540, Munich, Germany
First published on 12th December 2025
Abstract
Electrospun protein-based nanofibers offer a renewable and biocompatible alternative to fully synthetic materials, benefiting from the use of naturally derived components and reduced reliance on petrochemical polymers. Despite their promise, the relationship between processing conditions and fiber morphology remains poorly understood. Here, we present a systematic study of bovine serum albumin:polyethylene oxide (BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO) nanofibers, focusing on controlling morphology and functionalization for biosensing applications. Electrospinning parameters, solution composition, and pretreatment procedures were optimized to improve process stability and reproducibility of protein-based fibers with specific morphologies. To gain insight into the chemical composition of the fibers, we used advanced characterization techniques such as scattering-type scanning near-field optical microscopy (s-SNOM) with nano-FTIR spectroscopy. This, combined with two-photon-excited green autofluorescence exhibited by the proteins in electrospun fibers, allowed us to examine the internal architecture and provide evidence of molecular-scale structural repeatability. The optimized BSA
PEO) nanofibers, focusing on controlling morphology and functionalization for biosensing applications. Electrospinning parameters, solution composition, and pretreatment procedures were optimized to improve process stability and reproducibility of protein-based fibers with specific morphologies. To gain insight into the chemical composition of the fibers, we used advanced characterization techniques such as scattering-type scanning near-field optical microscopy (s-SNOM) with nano-FTIR spectroscopy. This, combined with two-photon-excited green autofluorescence exhibited by the proteins in electrospun fibers, allowed us to examine the internal architecture and provide evidence of molecular-scale structural repeatability. The optimized BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO fibers served as a biocatalytic layer in model electrochemical biosensors for dopamine detection, showing high sensitivity and reproducibility. These findings highlight protein–polymer composites as strong candidates for potential medical diagnostics, due to their renewable origin and functional versatility. The ability to tune morphology and investigate molecular structure opens new avenues for eco-friendly materials in healthcare and analytical science.
PEO fibers served as a biocatalytic layer in model electrochemical biosensors for dopamine detection, showing high sensitivity and reproducibility. These findings highlight protein–polymer composites as strong candidates for potential medical diagnostics, due to their renewable origin and functional versatility. The ability to tune morphology and investigate molecular structure opens new avenues for eco-friendly materials in healthcare and analytical science.
1. Introduction
Biopolymeric electrospun fibers, due to their biocompatibility, low toxicity, and biodegradability, have been successfully applied in a wide range of fields, including drug carriers,1 cell scaffolds for tissue engineering,2 functional medical dressings3 and photonics.4 The use of materials in the form of micro- and nanofibers in these areas is associated with a particularly high surface-to-volume ratio, ease of functionalization, and high porosity.5 Furthermore, the electrospinning technique is a relatively simple and cost-effective method that enables the production of fibers from naturally derived materials with customizable properties, including morphology, fiber diameter, spatial arrangement within the nonwoven structure, and fabric thickness, tailored to specific applications. Moreover, materials manufacturing can be scaled for industrial use, providing a relatively straightforward path for commercializing the developed products.6In the study of electrospinning biopolymers, special emphasis is placed on the simplicity and stability of the production process, material availability, and pretreatment requirements. Among naturally derived materials for electrospinning, bovine serum albumin (BSA) meets these requirements particularly effectively.7 BSA is a well-known, relatively inexpensive, neutral globular protein extracted on a large scale from cow blood.8 Its low reactivity, exceptional biocompatibility, and high resistance to denaturation have made BSA one of the best-characterized proteins, with a wide range of applications in biotechnology, including enzymatic stabilization, blocking agents, and as a model protein in biochemistry and clinical diagnostics.7 The extensive biophysical research and well-documented properties of BSA have also contributed to advancing knowledge about the influence of electrospinning process parameters on the resulting protein structures.
However, the electrospinning of globular proteins, such as BSA, into high-quality fibers is challenging. Two main approaches can be distinguished. The first one involves structural manipulation of the protein, leading to changes in protein conformation, aggregation, and the intra- and intermolecular exchange of disulfide bonds.8 Appropriate selection of solvents and reducing agents enables the transformation of a low-viscosity protein solution into a spinnable one.7 Furthermore, studies of Regev, O. et al.9 have shown that modifying protein conformation and concentration allows for controlling solution viscoelasticity, enabling the production of reproducible protein-based materials. A trifluoroethanol–water solvent mixture has been used to disrupt BSA's tertiary structure, induce partially folded conformations, and increase its hydrophobic surface.9,10 The addition of beta-mercaptoethanol aimed to reduce disulfide bonds, fully unfolding the protein. Whereas reduced BSA produced continuous, uniform fibers, untreated BSA led to fiber fragmentation.8
The second approach focuses on increasing the viscosity of the protein solution, which can be achieved by adding synthetic polymers as supporting agents.3 Biopolymers are typically blended with biocompatible, non-toxic, water-soluble polymers such as polyvinyl alcohol (PVA) and polyethylene oxide (PEO).11 Depending on the polymer concentration, these additives can have various functions, such as enhancing the mechanical properties of fiber mats and stabilizing the electrospinning process. For BSA, adding PEO dissolved in water also facilitated fiber formation due to the substantial increase in solution viscosity, which is a significant factor in the electrospinning process.12 Furthermore, adjusting the polymer-to-protein ratio allowed for the production of fibers with varying thicknesses.13
Nevertheless, both approaches are limited by certain drawbacks. In the first case, due to the denaturation process, some functionalities of proteins resulting from their higher-order structure might be lost. In the second one, the number of protein additives is rather limited, and thus their functional role might be suppressed by a dominating, spinnable polymer. Finally, a comprehensive and in-depth understanding of the electrospinability of proteins, in particular the globular ones, still remains to be clarified. Specifically, the relationship between fiber composition, protein processing parameters, and their effect on fiber morphology at the nanoscale remains incompletely understood, requiring further exploration and application of advanced microscopic techniques. In this context, we conducted a morphological investigation of nanofibers primarily based on BSA, used here as a model globular protein-based material in combination with PEO. To the best of our knowledge, this is the first such detailed analysis of the structure and composition of BSA-based electrospun materials, offering new insight into their nanoscale structure and organization.
In this research, electrospun fibers from BSA were fabricated using a hybrid approach that combined the assumptions of the two previously described methods. This strategy deviates from the conventional use of fully denatured proteins, which often compromises their functionality, and at the same time allows for increasing the protein content to as much as 90% of the fiber's total mass. A solvent mixture of water and dimethylformamide (DMF), in a volumetric ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, was used to partially unfold the protein, while PEO was added as a supporting polymer to increase viscosity. This combination enabled a highly stable process and reproducibility under a given set of conditions, allowing for the development of materials ranging from thin, single-fiber layers to thicker mats, for which BSA accounted for 90% of the total weight. This ratio not only promotes the material's biocompatibility and broad bio-related applications, but also facilitates the investigation of the structure of electrospun proteins.
3, was used to partially unfold the protein, while PEO was added as a supporting polymer to increase viscosity. This combination enabled a highly stable process and reproducibility under a given set of conditions, allowing for the development of materials ranging from thin, single-fiber layers to thicker mats, for which BSA accounted for 90% of the total weight. This ratio not only promotes the material's biocompatibility and broad bio-related applications, but also facilitates the investigation of the structure of electrospun proteins.
The produced material was comprehensively characterized with scattering-type scanning near-field optical microscopy (s-SNOM) with Fourier transform infrared nanospectroscopy (nano-FTIR) and transmission electron microscopy (TEM). The research focused on two morphological types of the material: uniform fibers with a uniform diameter and fibers formed through the phase non-homogeneity with the presence of beads. Moreover, the observed autofluorescence of electrospun BSA-based materials provided an opportunity to study the molecular orientation of BSA in electrospun fibers with polarization-sensitive two-photon microscopy. These studies indicate an ordered and repeatable internal structure of protein-based electrospun fibers.
To gain more insight into the structure of the BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO matrix and its functionality, we used fabricated fibers as a scaffold in two model electrochemical biosensors based on laccase for dopamine detection. In this case, the integration of a biocompatible fiber structure composed of BSA, commonly employed as an enzyme stabilizer,7 with a nanofibrous morphology that enhances the available surface area for analyte interaction, it fulfills the key criteria for biosensor applications. The fabricated platforms, featuring two distinct laccase immobilization strategies (on the surface and within the volume), were proposed and investigated. Both designs exhibited low detection thresholds, good stability, sensitivity, and selectivity, meeting the requirements necessary for detecting dopamine from biological samples. Finally, the successful fabrication of electrochemical dopamine sensors demonstrates that the hybrid approach used here preserves protein functionality and reduces the amount of auxiliary polymer to obtain fibers of stable morphologies.
PEO matrix and its functionality, we used fabricated fibers as a scaffold in two model electrochemical biosensors based on laccase for dopamine detection. In this case, the integration of a biocompatible fiber structure composed of BSA, commonly employed as an enzyme stabilizer,7 with a nanofibrous morphology that enhances the available surface area for analyte interaction, it fulfills the key criteria for biosensor applications. The fabricated platforms, featuring two distinct laccase immobilization strategies (on the surface and within the volume), were proposed and investigated. Both designs exhibited low detection thresholds, good stability, sensitivity, and selectivity, meeting the requirements necessary for detecting dopamine from biological samples. Finally, the successful fabrication of electrochemical dopamine sensors demonstrates that the hybrid approach used here preserves protein functionality and reduces the amount of auxiliary polymer to obtain fibers of stable morphologies.
2. Results and discussion
2.1 Electrospinning of BSA-based fibers
Solutions of BSA and PEO, dissolved in a solvent mixture of water and DMF in a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 volumetric ratio, resulted in different electrospun morphologies depending on the time elapsed after protein addition. In this case, the addition of DMF to the water was used to improve the electrospinning process, considering that it has lower surface tension than water, enabling easier jet formation at lower electric fields.14 Moreover, DMF can partially disrupt the BSA tertiary structure15 and generate partly unfolded conformations, which facilitate the process of continuous fiber formation.9 The use of DMF in the minority to water gives the desired results of producing a stable cone during the process and, consequently, the possible formation of thick electrospun fabrics (Fig. S1A, SI). Applying it in higher volumes resulted in apparent denaturation of the protein, which we wanted to avoid; therefore, a 7
3 volumetric ratio, resulted in different electrospun morphologies depending on the time elapsed after protein addition. In this case, the addition of DMF to the water was used to improve the electrospinning process, considering that it has lower surface tension than water, enabling easier jet formation at lower electric fields.14 Moreover, DMF can partially disrupt the BSA tertiary structure15 and generate partly unfolded conformations, which facilitate the process of continuous fiber formation.9 The use of DMF in the minority to water gives the desired results of producing a stable cone during the process and, consequently, the possible formation of thick electrospun fabrics (Fig. S1A, SI). Applying it in higher volumes resulted in apparent denaturation of the protein, which we wanted to avoid; therefore, a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 water
3 water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF ratio was established. We also tested solvents, such as DMSO and acetonitrile, in the same ratio as water
DMF ratio was established. We also tested solvents, such as DMSO and acetonitrile, in the same ratio as water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF; however, due to significant processing difficulties, we reverted to using DMF for this purpose (Fig. S1C–E, SI). It is worth noting that the solvent mixture is used to carry out the material fabrication process, but it evaporates during the procedure and is therefore not present in the final material.
DMF; however, due to significant processing difficulties, we reverted to using DMF for this purpose (Fig. S1C–E, SI). It is worth noting that the solvent mixture is used to carry out the material fabrication process, but it evaporates during the procedure and is therefore not present in the final material.
Electrospinning was performed as described in the Experimental section: 1.3 Electrospinning (SI). Within the first 3 hours after mixing protein with PEO solution, fibers with a characteristic bead structure were observed. However, keeping the same process conditions, after 4 hours of protein dissolution, the electrospinning resulted in regular fibers with a repeatable diameter. This trend was consistent across different protein and PEO concentrations. In contrast, electrospraying occurred when the PEO concentration was below 1%.
Fig. 1 presents the structures of fibers, obtained for a protein-to-polymer weight-to-weight ratio of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 after 2 and 4 hours after dissolution of BSA in the water–DMF–PEO solution. After 2 hours, two distinct types of beads were identified. The first type, hereafter referred to as type A (Fig. 1), is characterized by a regular circular shape with visible cut-offs, sequentially transitioning into a thin fiber. The average A-type bead diameter for a BSA
10 after 2 and 4 hours after dissolution of BSA in the water–DMF–PEO solution. After 2 hours, two distinct types of beads were identified. The first type, hereafter referred to as type A (Fig. 1), is characterized by a regular circular shape with visible cut-offs, sequentially transitioning into a thin fiber. The average A-type bead diameter for a BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO ratio of 90
PEO ratio of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 was 1.28 ± 0.17 µm. The second type, referred to as type B (Fig. 1), has a spindle-like structure with no distinct cut-off point, smoothly transitioning into the fiber. The widest diameter of these beads was around 610 ± 100 nm. These two specified bead types, A and B, appeared along the fibers without any apparent pattern, often interwoven (Fig. 1).
10 was 1.28 ± 0.17 µm. The second type, referred to as type B (Fig. 1), has a spindle-like structure with no distinct cut-off point, smoothly transitioning into the fiber. The widest diameter of these beads was around 610 ± 100 nm. These two specified bead types, A and B, appeared along the fibers without any apparent pattern, often interwoven (Fig. 1).
For the electrospun fibers obtained after 4 hours of protein dissolution, no beaded structures were observed. Instead, homogeneous fibers with a consistent diameter and wavy edges were formed, as shown in Fig. 1. The average fiber diameter for a BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO ratio of 90
PEO ratio of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 was 317 ± 24 nm.
10 was 317 ± 24 nm.
Despite variations in the protein-to-PEO ratio, the dependence of electrospun structure formation on dissolution time in the water–DMF–PEO solution remained unchanged. An increase in the proportion of PEO in the system resulted in slightly larger A-type bead and fiber diameters, whereas B-type beads exhibited relatively consistent values, as shown in Fig. 1. Longer protein dissolution and mixing times in the solvent mixture with PEO did not alter fiber morphology. Notably, these solutions remained suitable for electrospinning even after prolonged storage at room temperature, even after three months post-preparation, yielding fibers with the same morphology.
On this basis, it can be concluded that the slow dissolution of the protein in a water and DMF mixture resulted in non-homogeneity for up to 3 hours of stirring. When the BSA and PEO were well mixed over time and completely dissolved, materials with a bead-free fiber structure similar to that previously reported for BSA were obtained.12,13 The observed effect of this process is the formation of fibers with a reproducible bead size, comparable to those obtained for electrospun triblock copolymers SEBS in a THF/DMF solvent mixture.16
2.2 Nano-FTIR structural characteristics of two types of electrospun BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) PEO fibers
PEO fibers
FTIR-ATR analysis under bulk conditions reveals the general composition of protein-based fibers and confirms the proportion of BSA and PEO in the electrospun fibers (Section 3. FTIR-ATR spectroscopy for electrospun fibers, Fig. S2, (SI)). To gain more insight into the characteristics of beads and fibrous structures, the nano-FTIR s-SNOM technique was applied. This method has allowed for determining the local spectra of structures and observing the differences in contrast shown in the nano-FTIR absorption maps,17 which arise from the two-component structure of protein-based fibers. In particular, the study focused on assessing the reproducibility of the fiber structures and verifying the presence of distinct phases in the beaded fiber formations.
In the first step, a qualitative analysis was conducted on BSA-based fibers to verify their two-component structure. Nano-FTIR absorption maps were measured for two distinct wavenumbers: 1652 cm−1, characteristic of the amide I band in proteins,18 and 1111 cm−1, corresponding to PEO.19 As expected, the observed spectrum exhibited a strong amide I signal for both beaded and non-beaded fibers. For fibers without beads, the signal intensity was evenly distributed (Fig. 2A), whereas for the bead shown in Fig. 2C, high intensity is noticeable only in the bead.
Analyzing the PEO signal is more challenging since it constitutes only 10% (wt) of the tested samples. As a result, its signal on the IR maps is considerably weaker. Furthermore, thin native SiO2 oxide films (1–2 nm) can form due to air oxidation of the silicon wafer, with phonon absorption occurring in a similar IR range (1100–1200 cm−1). This may affect the contrast between the fiber and the substrate at 1111 cm−1. As it captures contributions from both the material and the oxides on the substrate, the background for this sample exhibits relatively high IR absorbance.
Furthermore, the observed signal is most intense at the fiber edges (Fig. 2A). This intensity is not related to a significantly increased PEO content; it originates from a background scattering effect associated with the sample's indirect illumination.20,21 The same edge effect of increased signal, especially on one edge of the electrospun fiber, was observed in s-SNOM nano-FTIR measurements of silk fibroin-polyurethane fibers. This phenomenon is particularly noticeable in complex samples with significant height differences.20
For fibers between the beads (Fig. 2C), the PEO signal is also very weak compared to the high background signal, whereas no signal for the protein was observed. The only exception is a very small thickening on the fiber, where the signal at 1111 cm−1 is intense (Fig. 2C). To gain more insight into small fibers between the beads, the FTIR scan area was significantly reduced to a 3 × 1 µm view.
AFM measurement result (Fig. 2B and Fig. S3 SI) shows that the fiber between the tested beads is only around 6 nm in height. The thin fibers between the beads and the thickening are mainly formed from PEO polymer, with absorption in this case, is similar to the oxide layer formed on the silicon wafer during the measurement. Moreover, a close-up view of the fiber between the beads (Fig. 2C) shows that the IR absorption of PEO and a small admixture of BSA in different parts of the thickening complements the totality, but does not overlap.
Next, we performed nano-FTIR spectral measurements to obtain a more precise compositional analysis of the sample at specific locations. Four spectral measurements were taken at different locations on the bead along the fiber and then averaged. The results, shown in Fig. 3, indicate that the bead is primarily composed of protein, with two intense peaks at 1659 cm−1 and 1547 cm−1. In the same way, spectra were also measured and analyzed for a very small fiber-forming structure, which, based on the results, corresponded to the signal for PEO with peaks at 1117 cm−1, 1148 cm−1, and 1061 cm−1.
To assess the homogeneity of the C-type fiber structure, a nano-FTIR absorption spectral linescan was performed across the fiber. A graphical representation of these measurements (25 spectra recorded across 1.25 µm) is shown in Fig. 4A, along with the averaged nano-FTIR absorption spectra. As observed in previous measurements, the amide I and amide II bands showed the highest absorption, while the intensity of the band corresponding to PEO is very weak.
Nevertheless, the signal intensities were reproducible for subsequent measuring points along the fiber. Based on these results, it can be concluded that the fiber has a homogeneous structure, with no apparent separation between protein and polymer, unlike fibers containing beads.
Furthermore, based on the comparison between the two types of electrospun material, there is also an apparent change in the PEO nano-FTIR spectra. For the crystalline form, according to the ref. 22 two closely spaced peaks are present at 1114 cm−1 and 1149 cm−1, while the band for the amorphous form is wider, with its maximum at 1109 cm−1. Nano-FTIR spectra showed similar IR absorption differences. For electrospun bead-less fibers, a nano-FTIR peak was observed at approximately 1110 cm−1 (Fig. S4, SI), which is associated with the amorphous form of PEO. In contrast, fiber thickening between the beads is characterized by three distinct peaks at 1117 cm−1, 1148 cm−1, and 1061 cm−1, indicating the crystalline form of PEO.
The differences in nano-FTIR spectra for different forms of PEO are similar to those previously described in the paper.23 These may indicate a more crystalline form in the case of incomplete dissolution of the polymer for fibers with beads, compared to homogeneous fibers, where the band for PEO corresponds to an amorphous form.
2.3 Secondary protein structure
Nano-FTIR spectral measurements allow for the conformational analysis of BSA in two distinct morphological structures of electrospun materials. The analysis focused on the spectral range of 1600–1700 cm−1, characteristic of the amide I band.18,24 Fig. 4 presents averaged spectra collected from different locations for two kinds of structures – along an electrospun bead-less fiber (Fig. 4B), as well as from a bead structure on the fiber (Fig. 4C). We performed a curve fitting with peak assignment to resolve overlapping IR absorption bands for protein characteristic secondary structures. Based on previously specified ranges,18,24–26 the nano-FTIR spectra after deconvolution give us the relative contribution of 4 components to a particular protein secondary structure for α-helix, β-turn, β-sheet, and β-antiparallel sheet.For the electrospun BSA-based fiber without beads, the contribution is attributed to α-helix structures, with a peak at 1654 cm−1, β-turns at 1673 cm−1, β-sheets at 1634 cm−1, and a smaller contribution from β-antiparallel sheets at 1694 cm−1. In the case of the bead structure, the highest peak also corresponds to α-helix structures at 1655 cm−1, followed by β-turns at 1675 cm−1, and minor contributions from β-sheets at 1634 cm−1 and β-antiparallel sheets at 1694 cm−1.
Based on the notable increase in β-sheet content in the protein within the bead-less fibers, which are formed after a longer dissolution time in the water/DMF solvent mixture. It can be concluded that prolonged dissolution leads to partial unfolding of the protein structure. In contrast, the higher contributions of α-helix and β-turn structures relative to β-sheets in the beaded fibers suggest that the secondary structure remains closer to that of native BSA. This may result from a shorter dissolution time, potentially preserving a partially crystalline form of the protein. Moreover, these results are consistent with the previously discussed study by Kumaran R. et al.15 on partial protein unfolding in the DMF solution.
2.4 Electrospinning-induced orientation of globular protein in fibrils
During the electrospinning process, the solution experiences high extensional forces due to its flow through the capillary system, as well as Coulombic forces during jet elongation and acceleration under the applied electric field.27 This is followed by a rapid solvent evaporation and solidification. Mentioned effects collectively can influence the molecular orientation of the individual components within the electrospinning solution.28 For electrospun synthetic polymers, molecular orientation has been successfully studied using techniques such as X-ray scattering29,30 and polarized Raman spectroscopy.31 In the case of electrospun protein-based fibers, orientation has been confirmed so far in amyloid-based fibers28 by polarized FTIR and anisotropic SAXS/WAXS scattering patterns. However, amyloids are well-known for their highly organized secondary protein structure,28 in contrast to the much less organized, small globular proteins, such as BSA. For this reason, we wanted to determine whether small proteins are molecularly oriented in a similar way within the fiber and whether protein localization in the fibers and beads can be assessed based on this.For BSA-based fibers, the observed protein green autofluorescence enables the study of fluorescence dipole orientation using polarization-sensitive two-photon microscopy to determine the alignment of protein molecules. For this purpose, the material was first examined spectroscopically, and then its alignment along the fiber's long axis was determined. This way, the arrangement of the protein itself was examined, as the polymer added, PEO, does not exhibit fluorescence.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and OH of subunits of the peptide backbone and different amino acid residues, which in turn facilitate effective through-space electronic communication among different groups with n and π-electrons.34 For BSA-based electrospun fibers, green autofluorescence has not been previously identified; however, the very high protein concentrations in the material suggest a similar mechanism for the observed fluorescence. Using fluorescence confocal microscopy, the emission of different types of structures observed in BSA-based fibers was investigated, along with the measurement of fluorescence lifetimes. Fig. 5A and B presents overlapped views for the material autofluorescence excited at 405 nm and the transmitted light image for more precise identification of individual structures. Fig. 5A shows both types of beads identified in Section 2.1: Electrospinning of BSA-based fibers, which apparently have increased fluorescence intensity compared to the fibers in between. This result corresponds to nano-FTIR measurements, evidencing the protein as the main building component of the beads and, thus, a factor in the occurrence of material autofluorescence. The apparent fluorescence of the fibers in Fig. 5B is much more homogeneous, with a slight enhancement at the fiber intersections.
O, and OH of subunits of the peptide backbone and different amino acid residues, which in turn facilitate effective through-space electronic communication among different groups with n and π-electrons.34 For BSA-based electrospun fibers, green autofluorescence has not been previously identified; however, the very high protein concentrations in the material suggest a similar mechanism for the observed fluorescence. Using fluorescence confocal microscopy, the emission of different types of structures observed in BSA-based fibers was investigated, along with the measurement of fluorescence lifetimes. Fig. 5A and B presents overlapped views for the material autofluorescence excited at 405 nm and the transmitted light image for more precise identification of individual structures. Fig. 5A shows both types of beads identified in Section 2.1: Electrospinning of BSA-based fibers, which apparently have increased fluorescence intensity compared to the fibers in between. This result corresponds to nano-FTIR measurements, evidencing the protein as the main building component of the beads and, thus, a factor in the occurrence of material autofluorescence. The apparent fluorescence of the fibers in Fig. 5B is much more homogeneous, with a slight enhancement at the fiber intersections.
 | ||
| Fig. 5 One-photon excited green autofluorescence of BSA-based fibers and exemplary polar graphs for two-photon excited autofluorescence. One-photon excited green autofluorescence: beaded fiber structure (A) and fiber without beads (B). Fluorescence lifetime: beaded fibers (C) and fibers without beads (D). Exemplary polar graphs for the location of electrospun beads-free fiber (E), a fiber between the beads (F) and a bead (G), dots represent experimental data for the angular dependence of two-photon excited fluorescence intensity collected at two orthogonal directions: X (black) and Y (blue), lines present the fitting of the angular dependence of the X and Y components of two-photon excited fluorescence intensity according to the model described in paper.35 | ||
Fluorescence lifetimes were measured sequentially for the different types of beads and fibers. Measurements were conducted for 415 nm excitation due to equipment limitations. The fluorescence decay for all structures was fitted with a monoexponential model (Table S1, SI). For A-type and B-type beads, the calculated fluorescent lifetime is 2.37 ± 0.25 ns and 2.36 ± 0.16 ns, respectively. For bead-free fibers, the value is 2.20 ± 0.22 ns and 2.18 ± 0.27 ns for fibers between the beads. These results indicate that although the fluorescence intensity for individual structures varies, the fluorescence lifetime itself for beads or fibers is very comparable.
The intensity of the two-photon excited autofluorescence distribution is presented in raster scans; Fig. S8A (SI) shows scans for a beads-free sample, and Fig. S8B for fibers with beads (SI). A notable contrast in autofluorescence intensity was observed between the beads and the fibers connecting them, which was also evident in the case of single-photon excitation (Fig. 5A).
Measurements reveal the molecular orientation within protein-based fibers, as evidenced by reproducible values of angle Ψ. Three exemplary polar graphs obtained for the measurement points are shown in Fig. 5E–G, one measured for the fiber without beads (Fig. 5E), the other for a fiber between the beads (Fig. 5F), and one for the bead structure (Fig. 5G). The mean value of Ψ for fibers is 28° with a mean ΔΨ = 12°, while in the case of beaded structures, the obtained data could not be reliably reproduced by the applied model,35,37 which indicates significantly less ordered structure. However, no significant differences in internal molecular orientation were observed between the fibers without beads or a fiber between the beads (Fig. 5F). On this basis, we can assume that, as in the case of charged polymers27,29,31 and amyloid structures,28 proteins are also molecularly oriented in the fiber structure.
2.5 Dopamine biosensor application
Due to the high surface-to-volume ratio and excellent nanoscale homogeneity of nanofibers, as demonstrated by nanospectroscopic methods, the fabricated C-type fibers were utilized as a scaffold in two model electrochemical biosensors to explore their stability and functional potential as a biocatalytic layer (Fig. 6). The BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO nanofibers (BSA
PEO nanofibers (BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO NFs) matrix provides a minimalistic yet highly effective platform for enzyme immobilization, eliminating the need for additional chemical linkers or multi-step surface modifications. BSA offers biocompatibility and binding affinity for biomolecules, while PEO ensures ionic conductivity and structural stability. This hybrid composition forms a permselective, electroactive environment that supports efficient enzyme function and reduces background interference.38–40 To validate the functional capacity of this material, we employed, for the first time to our knowledge, an electrochemical biosensor platform based on BSA
PEO NFs) matrix provides a minimalistic yet highly effective platform for enzyme immobilization, eliminating the need for additional chemical linkers or multi-step surface modifications. BSA offers biocompatibility and binding affinity for biomolecules, while PEO ensures ionic conductivity and structural stability. This hybrid composition forms a permselective, electroactive environment that supports efficient enzyme function and reduces background interference.38–40 To validate the functional capacity of this material, we employed, for the first time to our knowledge, an electrochemical biosensor platform based on BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO NFs to compare two enzyme immobilization strategies: surface binding versus internal entrapment (Fig. 7). Laccase (Lac) and dopamine (DA) were selected as the model enzyme and analyte, respectively. The sensing mechanism of dopamine was described in detail in SI (Section 9. Sensing mechanism of dopamine).
PEO NFs to compare two enzyme immobilization strategies: surface binding versus internal entrapment (Fig. 7). Laccase (Lac) and dopamine (DA) were selected as the model enzyme and analyte, respectively. The sensing mechanism of dopamine was described in detail in SI (Section 9. Sensing mechanism of dopamine).
Electrochemical measurement revealed that internal enzyme entrapment significantly outperformed surface immobilization. Cyclic voltammetry (CV) showed that laccase embedded within the BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO NFs yielded sharper dopamine oxidation peaks (∼0.22 V) and more defined redox features associated with the enzyme's T1 copper center, indicating improved electron transfer and stabilization within the nanostructured matrix.41,42 Differential pulse voltammetry (DPV) confirmed this trend, demonstrating a broad linear response (2.5–200 µM) with high correlation (R2 = 0.975), while surface-immobilized systems exhibited nonlinear behavior at intermediate concentrations (Fig. S9, SI). These findings underscore how the internal architecture of the nanofibers promotes uniform enzyme distribution, preserves catalytic function, and enhances analytical performance, making it a compelling strategy for developing reliable and scalable bioelectronic platforms. The sensor application serves here as a functional demonstration of material efficiency, with broader implications for interfacing soft biomaterials and redox-active systems.
PEO NFs yielded sharper dopamine oxidation peaks (∼0.22 V) and more defined redox features associated with the enzyme's T1 copper center, indicating improved electron transfer and stabilization within the nanostructured matrix.41,42 Differential pulse voltammetry (DPV) confirmed this trend, demonstrating a broad linear response (2.5–200 µM) with high correlation (R2 = 0.975), while surface-immobilized systems exhibited nonlinear behavior at intermediate concentrations (Fig. S9, SI). These findings underscore how the internal architecture of the nanofibers promotes uniform enzyme distribution, preserves catalytic function, and enhances analytical performance, making it a compelling strategy for developing reliable and scalable bioelectronic platforms. The sensor application serves here as a functional demonstration of material efficiency, with broader implications for interfacing soft biomaterials and redox-active systems.
The limit of detection (LOD) for the described bio-systems was calculated as shown in eqn (1):
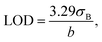 | (1) |
The detection limits for the biosensors were determined using eqn (1), yielding values of 0.0097 µM and 0.002 µM for laccase immobilized onto and inside the nanofibers, respectively. For context, typical dopamine concentrations in blood and urine range from 0 to 0.00025 µM and 0.3 to 3 µM, accordingly.43 The sensitivity of these biosensors was calculated as 4.6 and 3.2 µA mM−1 cm−2 for surface and internal immobilization, respectively. This range falls within the detection capabilities of the proposed biosensor.
To assess selectivity, the DPV method was used to evaluate the effect of potential interferents on dopamine signals. A 100 µM DA solution was spiked with 100 µM of interfering compounds, and the current responses were compared to DA alone. Fig. 7F shows negligible interference, with the system where laccase is entrapped within the nanofibers exhibiting the best selectivity (interference <8%). Both sensor configurations showed interference levels below 12%, confirming high selectivity.
Reproducibility was tested on six identical platinum electrodes (PtE) with a fixed 100 µM DA concentration. The surface-immobilized laccase system produced a relative standard deviation (RSD) of 2.2%, while the internal entrapment system demonstrated superior reproducibility with an RSD of 1.4%.
Stability was examined by storing the biosensors at 4 °C for two weeks. The system with laccase inside the BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO NFs retained nearly 100% of its initial signal, while the surface-immobilized system showed a 7% loss, likely due to enzyme detachment. When compared to a study on horseradish peroxidase immobilized in PVA:BSA nanofibers, which retained 73% of activity after 11 reaction cycles,44 the internal entrapment approach exhibited significantly higher stability and enzyme retention.
PEO NFs retained nearly 100% of its initial signal, while the surface-immobilized system showed a 7% loss, likely due to enzyme detachment. When compared to a study on horseradish peroxidase immobilized in PVA:BSA nanofibers, which retained 73% of activity after 11 reaction cycles,44 the internal entrapment approach exhibited significantly higher stability and enzyme retention.
The visibly enhanced performance observed for the system with laccase immobilized within the nanofibers can be attributed, firstly, to the creation of a more stable microenvironment that minimizes enzyme leaching during analysis, an issue more pronounced in surface-bound immobilization, and secondly, to the greater conformational flexibility afforded by internal entrapment. This internal orientation potentially exposes a higher number of active sites to the analyte, as the enzyme molecules can adopt diverse spatial arrangements rather than being confined to a uniform, planar distribution typical of surface immobilization.
Table 1 presents a comparative analysis of various electrochemical DA sensors utilizing nanofibers and other nanomaterials. It highlights key performance metrics such as sensitivity, selectivity, and detection limits, and demonstrates advancements in biosensor technology for neurotransmitter detection. Electrochemical methods are widely applied for DA detection. Various electrode materials, such as polymers, carbon nanostructures, metals, metal oxides, and their composites, have been developed to improve detection performance.45 In the current literature, most of the research is focused on carbon nanofibers and the immobilization strategy, which is based on anchoring the bioactive material onto the surface of nanofibers, not inside the nanofibers. In this case, the solvent mixture used for electrospinning the protein and PEO allowed the incorporation of the enzyme into the fiber, while retaining its catalytic activity, thereby enhancing the potential applicability of the resulting material.
| Type of electrode | LOD | Linear range | Sensitivity | Selectivity | Ref. |
|---|---|---|---|---|---|
| Biosensors utilizing nanofibers | |||||
| Co, Mo@CNFs/GCE | 0.00235 µM | 0.01–1000 µM | — | No effect towards K+, Na+, Cl−, NO3−, Cys, AA, GSH, Arg, Glc, Lys, Glu | 46 |
| FTO glass/PANI/f-CNTs NFs | 0.0974 µM | 0.005–0.5 µM | 6.88 µA cm−2 µM−1 | Selective in the presence Glu, UA, AA | 47 |
| Pd/CNF-CPE | 0.2 µM | 0.5–160 µM | n/d | Selective in the presence of AA, UA | 48 |
| Biosensors utilizing other nanomaterials | |||||
| GCE/CDs/Lac | 0.08 µM | 0.25–76.81 µM | n/d | Selective in the presence UA, AA, Na+, Ca2+, K+, Cl−, Gly, Glc, Ala | 49 |
| GCE/SiO2-PA/Lac | 0.26 µM | 0.99–103.10 µM | n/d | Sensitive in the presence UA, AA, Glc, Cys, phenol | 50 |
| LAC-Ni/Au/polipirol-COOH/NAF/SPE | 0.002265 µM | 0.01–50 µM | n/d | No effect towards AA, UA, Cys, Glc, 5-HT | 51 |
| Our study | |||||
Pt/BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO NFs/Lac (laccase inside the NFs) PEO NFs/Lac (laccase inside the NFs) |
0.002 µM | 2.5–200 µM | 3.2 µA mM−1 cm−2 | >8% towards AA, UA, urea, Tyr | This work |
3. Conclusions
This study presents a hybrid method for creating stable, protein-based nanofibers by partially unfolding globular proteins like BSA using a water:DMF solvent system, while reducing the need for auxiliary polymers such as PEO. The method ensures high process stability and reproducibility under specific conditions, allowing the production of materials with different structures, from thin fiber layers to thicker mats, with BSA making up to 90% of the total mass.Importantly, we demonstrate that fiber morphology can be precisely controlled by adjusting the dissolution time: shorter times (<3 hours) produce bead-containing fibers, while longer times result in uniform structures. This morphological tunability also affects the nanoscale chemical distribution within the fibers, as confirmed by s-SNOM with nano-FTIR spectroscopy. Additionally, the intrinsic green autofluorescence of BSA allows for non-invasive visualization of molecular organization inside the nanofibers, revealing a highly ordered and consistent internal structure. This level of morphological and molecular characterization is, to the best of our knowledge, highly novel for BSA-based electrospun materials and represents a significant advancement in understanding their nanoscale organization.
The significance of these materials was demonstrated through their use in electrochemical biosensors for dopamine detection. Two different methods of immobilizing laccase, inside the fiber matrix and on its surface, were tested. Enzyme immobilized inside the fibers produced clearer signals and had better long-term stability, compared to laccase immobilized on the surface of the nanofibers; however, both configurations demonstrated very good operational performance. These findings highlight the versatility of BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEO nanofibers as a sustainable and high-performance platform for biosensing. Moreover, the electrospinning process's scalability and compatibility with electrochemical biosensors and electronic and microfabrication technologies provide a strong basis for potential integration into miniaturized, real-time sensing platforms.
PEO nanofibers as a sustainable and high-performance platform for biosensing. Moreover, the electrospinning process's scalability and compatibility with electrochemical biosensors and electronic and microfabrication technologies provide a strong basis for potential integration into miniaturized, real-time sensing platforms.
Overall, this work enhances the design of protein-based nanomaterials by linking naturally derived components with biosensor applications, supporting their future use in diagnostics and bioanalytical technologies. In particular, by utilizing BSA obtained from animal by-products, this study highlights the potential of proteins for sustainable nanofiber fabrication.
Author contributions
K. K: conceptualization, investigation, data curation, formal analysis, validation, visualisation, writing – original draft, and writing – review & editing; S. B.: methodology, investigation, data curation, validation, formal analysis, writing – original draft; A. C.: methodology, investigation, data curation, validation, formal analysis, visualization and writing – review & editing; M. W.: methodology, investigation, data curation, and writing – review & editing; M. L.: investigation, formal analysis, validation, visualization; J. O.-B.: methodology, conceptualization, resources, and writing – review & editing; J. C.: resources, visualisation and writing – review & editing; L. S.: conceptualization, methodology, data curation, visualisation, writing – review & editing, resources, supervision, funding acquisition, and project administration.Conflicts of interest
The authors declare no conflict of interest.Data availability
Data for this article, including spectroscopic data (nano-FTIR, fluorescence spectra), imaging data (TEM, AFM), and electrochemical measurements, are available at RepOD Repository for Open Data (https://doi.org/10.18150/3QLUCA).The data supporting this article are also included as part of the supplementary information (SI), providing more comprehensive information to facilitate a deeper understanding of the study's findings. Supplementary information is available. See DOI: https://doi.org/10.1039/d5tb02207j.
Acknowledgements
The research was financed by the National Science Centre, Poland, grant no. 2021/41/B/ST5/01797. This publication was also partially developed under the provision of the Polish Ministry of Science and Higher Education project “Support for research and development with the use of research infrastructure of the National Synchrotron Radiation Centre SOLARIS” under contract no. 1/SOL/2021/2. We acknowledge the SOLARIS Centre for access to the CIRI beamline, where the measurements were performed. The authors thank M. Majkowski for fluorescent confocal microscopy with fluorescence lifetime imaging and the Attocube company for the possibility of the Nano-FTIR measurements. TOC, Fig. 6, and 7 were created using https://BioRender.com.References
- S. Zhao, C. Huang, X. Yue, X. Li, P. Zhou, A. Wu, C. Chen, Y. Qu and C. Zhang, Mater. Des., 2022, 110850, DOI:10.1016/j.matdes.2022.110850.
- M. D. Popov Pereira da Cunha, P. C. Caracciolo and G. A. Abraham, Curr. Opin. Biomed. Eng., 2021, 100243, DOI:10.1016/j.cobme.2020.07.003.
- J. Xue, T. Wu, Y. Dai and Y. Xia, Chem. Rev., 2019, 5298–5415, DOI:10.1021/acs.chemrev.8b00593.
- X. Yang, L. Xu, S. Xiong, H. Rao, F. Tan, J. Yan, Y. Bao, A. Albanese, A. Camposeo, D. Pisignano and B. Li, Nano Lett., 2024, 24, 566–575 Search PubMed.
- S. Deshmukh, M. Kathiresan and M. A. Kulandainathan, Biomater. Sci., 2022, 4424–4442, 10.1039/d2bm00820c.
- D. Ji, Y. Lin, X. Guo, B. Ramasubramanian, R. Wang, N. Radacsi, R. Jose, X. Qin and S. Ramakrishna, Nat. Rev. Methods Primers, 2024, 4(1), 1–21, DOI:10.1038/s43586-023-00278-z.
- M. Hayat, S. A. R. Bukhari and M. Irfan, Challenges and Future Directions, Biotechnol. J., 2023, 18(12), e23000279, DOI:10.1002/biot.202300279.
- Y. Dror, T. Ziv, V. Makarov, H. Wolf, A. Admon and E. Zussman, Biomacromolecules, 2008, 9, 2749–2754, DOI:10.1021/bm8005243.
- O. Regev, S. Vandebril, E. Zussman and C. Clasen, Polymer, 2010, 51(12), 2611–2620, DOI:10.1016/j.polymer.2010.03.061.
- R. Carrotta, M. Manno, F. M. Giordano, A. Longo, G. Portale, V. Martorana and P. L. San Biagio, Phys. Chem. Chem. Phys., 2009, 11, 4007–4018 Search PubMed.
- J. D. Schiffman and C. L. Schauer, Polym. Rev., 2008, 317–352, DOI:10.1080/15583720802022182.
- T. Kowalczyk, A. Nowicka, D. Elbaum and T. A. Kowalewski, Biomacromolecules, 2008, 9, 2087–2090 Search PubMed.
- R. Díaz-Puertas, E. Rodríguez-Cañas, M. J. Lozoya-Agulló, P. V. Badía-Hernández, F. J. Álvarez-Martínez, A. Falcó and R. Mallavia, Int. J. Biol. Macromol., 2024, 280, 136019, DOI:10.1016/j.ijbiomac.2024.136019.
- B. Honti, B. Fábián, A. Idrissi and P. Jedlovszky, J. Phys. Chem. B, 2023, 127, 1050–1062 Search PubMed.
- R. Kumaran and P. Ramamurthy, J. Lumin., 2014, 148, 277–284 Search PubMed.
- L. Wang, P. D. Topham, O. O. Mykhaylyk, H. Yu, A. J. Ryan, J. P. A. Fairclough and W. Bras, Macromol. Rapid Commun., 2015, 36, 1437–1443 Search PubMed.
- D. Eliaz, S. Paul, D. Benyamin, A. Cernescu, S. R. Cohen, I. Rosenhek-Goldian, O. Brookstein, M. E. Miali, A. Solomonov, M. Greenblatt, Y. Levy, U. Raviv, A. Barth and U. Shimanovich, Nat. Commun., 2022, 13, 7856, DOI:10.1038/s41467-022-35505-w.
- A. Barth, Biochim. Biophys. Acta, Bioenerg., 2007, 1073–1101, DOI:10.1016/j.bbabio.2007.06.004.
- I. Pucić and T. Jurkin, Radiat. Phys. Chem., 2012, 81, 1426–1429 Search PubMed.
- L. Mester, A. A. Govyadinov and R. Hillenbrand, Nanophotonics, 2022, 11, 377–390 Search PubMed.
- A. Caldiroli, S. Cappelletti, G. Birarda, A. Redaelli, S. Adele Riboldi, C. Stani, L. Vaccari and F. Piccirilli, Analyst, 2023, 148, 3584–3593 Search PubMed.
- C. Bergeron, E. Perrier, A. Potier and G. Delmas, Int. J. Spectrosc., 2012, 1–13 Search PubMed.
- O. David, M. Etxeberria Benavides, I. Amenabar Altuna, F. J. Fernandez Carretero, M. D. M. Diaz De Guereñu Zabarte, J. J. Flat, Q. Pineau, M. Goikoetxea Larruskain and R. Hillenbrand, J. Membr. Sci., 2024, 708, 123001, DOI:10.1016/j.memsci.2024.123001.
- H. Yang, S. Yang, J. Kong, A. Dong and S. Yu, Nat. Protoc., 2015, 10, 382–396 Search PubMed.
- H. A. Alhazmi, M. S. Alam, M. Albratty, A. Najmi, A. A. Abdulhaq, R. Hassani, W. Ahsan and A. N. Qramish, J. Chem., 2023, 1, 581653, DOI:10.1155/2023/2581653.
- A. Dong, P. Huang and W. S. Caughey, Protein Secondary Structures in Water from Second-Derivative Amide I Infrared Spectra, 1990, vol. 29 Search PubMed.
- T. Kongkhlang, K. Tashiro, M. Kotaki and S. Chirachanchai, J. Am. Chem. Soc., 2008, 130, 15460–15466 Search PubMed.
- D. Chen, N. Narayanan, E. Federici, Z. Yang, X. Zuo, J. Gao, F. Fang, M. Deng, O. H. Campanella and O. G. Jones, Biomacromolecules, 2020, 21, 2772–2785 Search PubMed.
- J. Liu, D. Y. Lin, B. Wei and D. C. Martin, Polymer, 2017, 118, 143–149 Search PubMed.
- L. Gong, D. B. Chase, I. Noda, J. Liu, D. C. Martin, C. Ni and J. F. Rabolt, Macromolecules, 2015, 48, 6197–6205 Search PubMed.
- L. M. Bellan and H. G. Craighead, Polymer, 2008, 49, 3125–3129 Search PubMed.
- X. Ma, T. Wang, D. Song, D. Hargrove, Q. Dong, Z. Luo, J. Chen, X. Lu, Y. Luo, T. H. Fan and Y. Lei, ACS Biomater. Sci. Eng., 2016, 2, 954–962 Search PubMed.
- L. Liu, G. Li, N. Xiang, X. Huang and K. Shiba, Sensors, 2020, 118, 143–149, DOI:10.3390/s20205886.
- Q. Wang, X. Dou, X. Chen, Z. Zhao, S. Wang, Y. Wang, K. Sui, Y. Tan, Y. Gong, Y. Zhang and W. Z. Yuan, Angew. Chem., 2019, 131, 12797–12803 Search PubMed.
- P. Obstarczyk, M. Lipok, M. Grelich-Mucha, M. Samoć and J. Olesiak-Bańska, J. Phys. Chem. Lett., 2021, 12, 1432–1437 Search PubMed.
- P. Obstarczyk, M. Lipok, A. Żak, P. Cwynar and J. Olesiak-Bańska, Biomater. Sci., 2022, 10, 1554–1561 Search PubMed.
- M. Lipok, P. Obstarczyk and J. Olesiak-Bańska, J. Visualized Exp., 2023,(199), 1–21, DOI:10.3791/65670.
- S. Pardhiya, U. S. Gaharwar, A. M. Parambil, J. P. Nirala and P. Rajamani, Free Radical Res., 2024, 58(3), 194–216 Search PubMed.
- C. O. Saeed, A. A. Qader and S. B. Aziz, Opt. Mater., 2022, 129, 112502, DOI:10.1016/j.optmat.2022.112502.
- S. Malinowski, M. Wardak and C. Wardak, Langmuir, 2024, 40(7), 3472–3485, DOI:10.1021/acs.langmuir.3c02942.
- H. Xu, L. Wang, J. Luo, Y. Song, J. Liu, S. Zhang and X. Cai, Sensors, 2015, 15, 1008–1021 Search PubMed.
- C. Adam, P. Scodeller, M. Grattieri, M. Villalba and E. J. Calvo, Bioelectrochemistry, 2016, 109, 101–107 Search PubMed.
- F. Kurniawan, N. S. Al Kiswiyah, K. A. Madurani and M. Tominaga, ECS J. Solid State Sci. Technol., 2017, 6, M3109–M3112 Search PubMed.
- R. Fazel, S. F. Torabi, P. Naseri-Nosar, S. Ghasempur, S. O. Ranaei-Siadat and K. Khajeh, Enzyme Microb. Technol., 2016, 93–94, 1–10 Search PubMed.
- M. Nie, S. Lu, D. Lei, C. Yang and Z. Zhao, J. Electrochem. Soc., 2017, 164, H952–H957 Search PubMed.
- Y. Xing, C. Lv, Y. Fu, L. Luo, J. Liu, X. Xie and F. Chen, Talanta, 2024, 271, 125674, DOI:10.1016/j.talanta.2024.125674.
- C. Kaewda and S. Sriwichai, Biosensors, 2024, 14(7), 349, DOI:10.3390/bios14070349.
- J. Huang, Y. Liu, H. Hou and T. You, Biosens. Bioelectron., 2008, 24, 632–637 Search PubMed.
- R. Wu, S. Yu, S. Chen, Y. Dang, S. H. Wen, J. Tang, Y. Zhou and J. J. Zhu, Anal. Chim. Acta, 2022, 1229, 340365, DOI:10.1016/j.aca.2022.340365.
- K. Wang, P. Liu, Y. Ye, J. Li, W. Zhao and X. Huang, Sens. Actuators, B, 2014, 197, 292–299 Search PubMed.
- S. Evli and D. Aktaş Uygun, Electroanalysis, 2023, 35(8), e202300012, DOI:10.1002/elan.202300012.
| This journal is © The Royal Society of Chemistry 2026 |






