Open Access Article
Open Access ArticleEngineering viral vectors with CD200 enhances anti-inflammatory and phagocytosis resistance
Esmael M. Alyami†
a,
Ian Pengb,
Sharjeel Jokhioa and
Ching-An Peng *a
*a
aDepartment of Chemical and Biological Engineering, University of Idaho, 875 Perimeter Drive MS 0904, Moscow, ID 83844, USA. E-mail: capeng@uidaho.edu; Tel: +1 208-885-7461
bDepartment of Bioengineering, University of Pennsylvania, Philadelphia, PA 19104, USA
First published on 22nd December 2025
Abstract
CD200, an immunoregulatory glycoprotein of the immunoglobulin superfamily, suppresses inflammatory signaling by engaging its receptor CD200R, which is predominantly expressed on myeloid cells. To enhance the immune-evading properties of viral vectors, we engineered lentiviral particles displaying the CD200 ectodomain (CD200ED) to exploit anti-inflammatory response and phagocytosis resistance. A fusion gene encoding the mouse CD200 ectodomain and core streptavidin (CD200ED-coreSA) was cloned into the pET-30a(+) plasmid, expressed in E. coli Lemo21(DE3), and purified via immobilized metal affinity chromatography (IMAC). Successful protein assembly was confirmed by SDS-PAGE and western blot. Biotinylated VSV-G pseudotyped lentiviral vectors, encoding a green fluorescent protein reporter, were functionalized with CD200ED-coreSA. When exposed to murine J774A.1 macrophages, CD200ED-modified lentiviruses significantly reduced pro-inflammatory cytokine production - evidenced by 47.1% decrease in TNF-α and 55% decrease in IL-6 - compared to unmodified controls. Additionally, CD200ED anchoring reduced macrophage phagocytosis of lentiviral particles by 25%. These findings demonstrate that CD200-tethering confers dual anti-inflammatory and phagocytosis resistance capabilities to viral vectors, offering a promising strategy to improve gene delivery efficiency in inflammatory environments.
Introduction
Viral therapeutics face significant challenges due to host innate immune defenses, including the production of inflammatory cytokines (e.g., TNF-α, IL-6), complement activation, and neutralizing antibodies, which collectively diminish systemic antitumor efficacy.1–6 Compounding this issue, intravenous delivery is hindered by rapid clearance via the reticuloendothelial system (RES), where liver-resident macrophages sequester up to 90% of viral particles within minutes of administration.7,8 To circumvent immune detection, innovative strategies have emerged to cloak viral vectors from immune scrutiny. For example, mesenchymal stem cells (MSCs) can act as immunologically inert carriers, shielding therapeutic viruses en route to tumor sites.9,10 Alternatively, chemical camouflage – such as polyethylene glycol (PEG) conjugation to viral surface proteins – masks epitopes to evade immunosurveillance,11,12 though this approach risks reduced infectivity and anti-PEG immune reactions.13,14 A biomimetic strategy inspired by endogenous immune checkpoints involves CD47, an immunoglobulin superfamily glycoprotein that signals “self” by binding macrophage SIRPα receptors to inhibit phagocytosis.15–17 Building on this mechanism, studies have engineered CD47-functionalized lentiviral vectors, leveraging the CD47-SIRPα axis to prolong circulation and enhance delivery efficiency.18–20CD200, a transmembrane glycoprotein of the immunoglobulin superfamily, functions as a key modulator of inflammatory responses and is broadly expressed across diverse cell types, including myeloid cells (macrophages, dendritic cells), lymphoid cells (T and B cells), neurons, keratinocytes, and endothelial cells.21–25 Its extracellular domain comprises two conserved regions: an N-terminal immunoglobulin-variable (IgV) domain and a membrane-proximal immunoglobulin-constant (IgC) domain, which mediate ligand-receptor interactions.26 CD200 binds exclusively to its cognate receptor, CD200R, a transmembrane protein expressed predominantly on myeloid cells (e.g., macrophages, neutrophils, dendritic cells, microglia)27–29 and subsets of lymphoid cells.28,29 The CD200–CD200R axis delivers potent immunosuppressive signals that inhibit macrophage activation and inflammatory cytokine production.27
In cancer biology, CD200 expression on cancer stem cells (CSCs) facilitates immune evasion by suppressing myeloid-derived suppressor cells (MDSCs) and dendritic cell maturation, enabling tumor persistence.30 This inhibitory pathway also dampens pro-inflammatory responses in autoimmune and neurodegenerative diseases.31–33 Mechanistically, CD200–CD200R engagement attenuates Toll-like receptor (TLR)-driven immune activation, limiting the secretion of cytokines such as IL-1β, IL-6, and TNF-α.34,35 Notably, dysregulation of this axis is implicated in chronic inflammation, making it a promising therapeutic target for conditions like multiple sclerosis, rheumatoid arthritis, and Alzheimer's disease.36–39
While the CD200–CD200R axis is well-characterized for its suppression of pro-inflammatory cytokines (e.g., TNF-α, IL-6) in myeloid cells, emerging evidence highlights its dual role in modulating phagocytic activity. For instance, CD200-Fc fusion protein administration in vivo enhances white matter repair by inhibiting phagocytosis of oligodendrocyte precursors, suggesting therapeutic potential in demyelinating diseases.40 Conversely, microglia isolated from CD200−/− mice exhibit hyperphagocytic behavior, engulfing amyloid-β aggregates and synthetic particles at elevated rates.41 This phenomenon extends to engineered systems: nanoparticles functionalized with the CD200 ectodomain (IgV + IgC regions) reduce phagocytosis by human THP-1 macrophages by downregulating surface TLR4 expression.42 Moreover, the secretion of pro-inflammatory cytokine tumor necrosis factor-α (TNF-α) from THP-1 macrophages treated with CD200-coated particles was diminished.42 Mechanistically, CD200 binding to CD200R triggers Src-family kinase-mediated phosphorylation of the receptor's intracellular domain, recruiting adaptor protein downstream of tyrosine kinase 2 (Dok2) to suppress mitogen activated protein kinase (MAPK) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling - pathways critical for both phagocytosis and inflammatory responses.43–45 Dok2 further intersects with TLR4 regulation, creating a feedback loop that amplifies immune tolerance.46
Notably, pathogens have evolutionarily co-opted this checkpoint; herpesviruses47 and poxviruses48 encode CD200 homologs to evade host immunity, underscoring its potency as an immunosuppressive tool. Inspired by these mechanisms, we engineered lentiviral vectors displaying the CD200 IgV domain to mimic endogenous immune evasion. Our results demonstrate that CD200-tethering concurrently attenuates pro-inflammatory cytokine release and macrophage phagocytosis, potentially prolonging viral persistence in circulation. This biomimetic strategy holds promise for improving gene delivery in inflammatory milieus, such as tumors or autoimmune environments.
Materials and methods
Plasmid construction for CD200ED-coreSA fusion
The plasmid encoding the mouse CD200 ectodomain-core streptavidin (mCD200ED-coreSA) fusion protein was constructed through sequential cloning steps. Core streptavidin (coreSA) was amplified via PCR from plasmid pSTE2-215 (yol)49 using primers flanked by EcoRI and XhoI restriction sites (forward: 5′-AGATCCGAATTCGGTGCTGCTGAAGCAGGT-3′; reverse: 5′-ATTATACTCGAGGGAGGCGGCGGACGGCTT-3′). Separately, the IgV ectodomain of mouse CD200 (CD200ED) was amplified from a pCMV-SPORT6 clone (Harvard Medical School) with primers containing BamHI and EcoRV sites (forward: 5′- GCGATGGCCATGGATATCATGTGGCAGAAAAAGAAAGCC-3′; reverse: 5′-GCTCGAATTCGGATCCGCAAGTGATGTTTAGGTGGTC-3′). PCR reactions (50 µL) consisted of 1× Phusion High-Fidelity Buffer, 200 µM dNTPs, 0.5 µM primers, 1 µg template DNA, and 1 U Phusion DNA Polymerase (New England Biolabs). Amplification conditions included an initial denaturation at 98 °C for 30 s, followed by 30 cycles of 98 °C (10 s), 60 °C (15 s), and 72 °C (15 s), with a final extension at 72 °C for 5 min. PCR products were purified using a Monarch PCR DNA Cleanup Kit (NEB), resolved on a 1% agarose gel, and digested with their respective restriction enzymes. The coreSA fragment was cloned into pET-30a(+) via EcoRI/XhoI to generate plasmid pME005. Subsequently, CD200ED was inserted into pME005 using BamHI/EcoRV, yielding the final construct pME007 (Fig. 1A), which was sequence-verified (Genewiz).Recombinant expression and purification of CD200ED-coreSA fusion protein
The CD200ED-coreSA fusion protein was expressed and purified using a modified protocol from our prior work.50 Briefly, the pME007 plasmid was transformed into E. coli Lemo21(DE3) cells, plated on lysogeny broth (LB) agar containing 50 µg mL−1 kanamycin, and incubated overnight at 37 °C. A single colony was inoculated into 5 mL LB-kanamycin medium and grown overnight at 37 °C with shaking (230 rpm). This starter culture was transferred to 1 L LB-kanamycin medium supplemented with 500 µM L-rhamnose (to regulate T7 RNA polymerase expression) and incubated at 37 °C (225 rpm) until reaching an OD600 of 0.6–0.8. Protein expression was induced with 400 µM isopropyl β-D-1-thiogalactopyranoside (IPTG), followed by overnight incubation at 18 °C. Cells were harvested by centrifugation (4500×g, 20 min), resuspended in B-PER bacterial protein extraction reagent (5 mL g−1 pellet) containing 50 mM Tris-HCl (pH 7.5) and 1× EDTA-free protease inhibitor, and incubated at room temperature for 20 min. Lysates were sonicated on ice (Misonix Sonicator 3000, 5 W output, 30 min total with 10 s pulses/20 s cooling intervals) and clarified by centrifugation (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 20 min).
000×g, 20 min).
The CD200ED-coreSA fusion protein was purified via immobilized metal affinity chromatography (IMAC) using HisPur™ Co-NTA resin. The resin was equilibrated with two bed volumes of binding buffer (10 mM imidazole in PBS, pH 7.4), after which the clarified lysate was mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with binding buffer and incubated with the resin for 2 hours at 4 °C to enhance histidine-tagged protein binding. Unbound proteins were removed by washing with equilibration buffer until the UV280 absorbance stabilized at baseline. Bound CD200ED-coreSA was eluted using 250 mM imidazole in PBS (pH 7.4), and fractions were pooled and concentrated using a 10 kDa molecular weight cutoff (MWCO) polyethersulfone centrifugal filter (Pierce). Protein concentration was quantified via BCA assay, while purity and identity were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot under reducing conditions using antibodies (detailed below). For comparative analysis, the coreSA control protein (encoded by plasmid pME005) was expressed and purified identically.
1 with binding buffer and incubated with the resin for 2 hours at 4 °C to enhance histidine-tagged protein binding. Unbound proteins were removed by washing with equilibration buffer until the UV280 absorbance stabilized at baseline. Bound CD200ED-coreSA was eluted using 250 mM imidazole in PBS (pH 7.4), and fractions were pooled and concentrated using a 10 kDa molecular weight cutoff (MWCO) polyethersulfone centrifugal filter (Pierce). Protein concentration was quantified via BCA assay, while purity and identity were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot under reducing conditions using antibodies (detailed below). For comparative analysis, the coreSA control protein (encoded by plasmid pME005) was expressed and purified identically.
SDS-PAGE and western blot analysis
SDS-PAGE was conducted using 12% polyacrylamide Mini-PROTEAN Tetra handcast gels to analyze both the soluble crude protein and the purified CD200ED-coreSA recombinant protein. Prior to electrophoresis, samples were mixed with an equal volume of 2× Laemmli sample buffer containing 5% (v/v) 2-mercaptoethanol and heated at 95 °C for 5 minutes. Electrophoresis was performed using tris-glycine electrode buffer (1×, pH 8.3) at 200 V for 40 minutes. The gel was stained with Coomassie Brilliant Blue R250 staining solution, which contained 40% (v/v) methanol and 10% (v/v) acetic acid, for a minimum of 3 hours. Following staining, the gel was destained in two steps: first, it was fast destained with a solution containing 40% (v/v) methanol and 10% (v/v) acetic acid for 1 hour at room temperature, and then it was incubated in a slow-destaining solution containing 10% (v/v) methanol and 10% (v/v) acetic acid overnight.For western blot analysis, three electrophoresed samples from the SDS-PAGE were transferred onto a 0.45 µm PVDF membrane using a Trans-Blot® semi-dry transfer system (Bio-Rad) at 20 V for 1 hour. The blots were then blocked with blocking buffer (5% bovine serum albumin (BSA) in tris-buffered saline with 0.1% Tween 20 (TBST)) for 1 hour at room temperature. Following blocking, the blots were washed three times with TBST and incubated separately with the following primary antibodies: anti-mouse CD200 monoclonal antibody, anti-streptavidin monoclonal antibody, and anti-histag monoclonal antibody. Incubation was performed under gentle shaking at 4 °C overnight. After washing the blots three times with TBST, they were separately incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of horseradish peroxidase (HRP)-labeled mouse IgG (secondary antibody) at room temperature for 2 hours. Following another three washes with TBST, the blots were developed using an ECL substrate and imaged using a chemiluminescence imager (PXi Syngene, Frederick, MD).
1000 dilution of horseradish peroxidase (HRP)-labeled mouse IgG (secondary antibody) at room temperature for 2 hours. Following another three washes with TBST, the blots were developed using an ECL substrate and imaged using a chemiluminescence imager (PXi Syngene, Frederick, MD).
Preparation of lentivirus and biotinylated lentivirus
HEK 293T cells were cultured in T-75 flasks in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin at 37 °C under a 5% CO2 atmosphere. To enable parallel production, the medium in one flask was additionally supplemented with 0.02 mg mL−1 Biotin-Cap-PE (Avanti Polar Lipids) for generating biotinylated lentivirus. For transfection, PEI/DNA complexes were prepared at an N/P ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A filter-sterilized PEI stock solution (200 µg mL−1, pH 7.0) was mixed with 6 µg of total plasmid DNA, comprising the packaging plasmid pCMV-dR8.2 (Addgene #12263), the envelope plasmid pMD2.G (Addgene #12259), and the reporter plasmid pLJM1-EGFP (Addgene #19319). The mixture was incubated at room temperature for 30 minutes to allow complex formation.
1. A filter-sterilized PEI stock solution (200 µg mL−1, pH 7.0) was mixed with 6 µg of total plasmid DNA, comprising the packaging plasmid pCMV-dR8.2 (Addgene #12263), the envelope plasmid pMD2.G (Addgene #12259), and the reporter plasmid pLJM1-EGFP (Addgene #19319). The mixture was incubated at room temperature for 30 minutes to allow complex formation.
Upon reaching 80% confluency, cells in both flasks were transfected with the prepared PEI/DNA complexes. Starting 24 hours post-transfection, viral supernatant was harvested daily and immediately replaced with fresh medium (with or without biotin–lipid supplement). The collected supernatants, corresponding to native and biotinylated lentivirus, were separately filtered through a 0.45 µm syringe filter and stored at −80 °C. The lentivirus harvested on the third day was concentrated using a Polybrene-based co-precipitation method.20 Briefly, the supernatant was mixed with an equal volume of polybrene solution (320 µg mL−1) and incubated at 37 °C for 30 minutes. The virus was then pelleted by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10 minutes at 4 °C. The final pellet was resuspended in 1 mL of phosphate-buffered saline (PBS) and stored at −80 °C for subsequent use.
000×g for 10 minutes at 4 °C. The final pellet was resuspended in 1 mL of phosphate-buffered saline (PBS) and stored at −80 °C for subsequent use.
Characterization of lentivirus and functionalized lentivirus
To distinguish between biotinylated and native lentiviruses concentrated using Polybrene, each sample was individually incubated with streptavidin-conjugated gold nanoparticles (SA-AuNPs, 10 nm in diameter, Nanocs) and prepared for transmission electron microscopy (TEM) analysis. Briefly, 10 µL of either biotinylated or native lentivirus was diluted in 1 mL of PBS containing 0.05 mg mL−1 SA-AuNPs and incubated at 4 °C for 1 hour with gentle shaking. Unbound gold nanoparticles were removed by two rounds of washing and centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10 minutes using 1× PBS. To prepare samples for TEM, 4 µL of the lentivirus-gold nanoparticle mixture was applied to a Formvar-coated grid (200-mesh size; Ted Pella, Redding, CA) and incubated under a fume hood for 2 minutes. The samples were then negatively stained with 4 µL of 2% phosphotungstic acid aqueous solution for 2 minutes and dried under a heat lamp for 30 minutes before imaging to visualize the lentiviral vectors and any attached gold nanoparticles. The images were taken at 200 kV (Tecnai G2 20 Twin; FEI Company, Hillsboro, OR). Dynamic light scattering and zeta potential analysis (Brookhaven Instruments Corporation, Holtsville, NY) of lentivirus, biotinylated lentivirus, and CD200ED-capped lentivirus were conducted to determine the median size and charge of different types of viruses.
000×g for 10 minutes using 1× PBS. To prepare samples for TEM, 4 µL of the lentivirus-gold nanoparticle mixture was applied to a Formvar-coated grid (200-mesh size; Ted Pella, Redding, CA) and incubated under a fume hood for 2 minutes. The samples were then negatively stained with 4 µL of 2% phosphotungstic acid aqueous solution for 2 minutes and dried under a heat lamp for 30 minutes before imaging to visualize the lentiviral vectors and any attached gold nanoparticles. The images were taken at 200 kV (Tecnai G2 20 Twin; FEI Company, Hillsboro, OR). Dynamic light scattering and zeta potential analysis (Brookhaven Instruments Corporation, Holtsville, NY) of lentivirus, biotinylated lentivirus, and CD200ED-capped lentivirus were conducted to determine the median size and charge of different types of viruses.
Immobilization of CD200ED-coreSA protein on biotinylated lentiviral vectors
CD200ED-coreSA was immobilized on the surface of biotinylated lentiviral vectors via specific biotin–streptavidin interactions. Briefly, 500 µL of biotinylated virus was mixed with 1 mL of CD200ED-coreSA protein in a microcentrifuge tube and gently rotated for 1 hour at 4 °C. Unbound CD200ED-coreSA protein was removed by two rounds of centrifugation and washing, and the pellet was resuspended in PBS. The successful attachment of CD200ED-coreSA to the viral surface was confirmed by dot blot assay. Specifically, 5 µL of the re-suspended sample was spotted onto a PVDF membrane, which was then blocked with blocking buffer for 1 hour at room temperature. The membrane was subsequently incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution of anti-mouse CD200 primary antibody (Santa Cruz, CA) in 1× TBST for 1 hour at room temperature. After three washes with TBST (5 minutes each), the membrane was incubated with a 1
500 dilution of anti-mouse CD200 primary antibody (Santa Cruz, CA) in 1× TBST for 1 hour at room temperature. After three washes with TBST (5 minutes each), the membrane was incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of anti-mouse IgG HRP-conjugated secondary antibody for 1 hour at room temperature. Following three additional TBST washes, HRP activity was detected to confirm the presence of CD200ED on the membrane. Biotinylated virus alone and soluble CD200ED-coreSA protein were used as negative and positive controls, respectively.
1000 dilution of anti-mouse IgG HRP-conjugated secondary antibody for 1 hour at room temperature. Following three additional TBST washes, HRP activity was detected to confirm the presence of CD200ED on the membrane. Biotinylated virus alone and soluble CD200ED-coreSA protein were used as negative and positive controls, respectively.
Effect of CD200ED-coreSA on pro-inflammatory cytokine secretion by macrophages
The effect of CD200ED-coreSA-tagged lentiviral particles on pro-inflammatory cytokine secretion was evaluated by measuring IL-6 and TNF-α levels in J774A.1 macrophages. J774A.1 cells (5 × 104) were cultured and treated with one of the following for 18 hours: unmodified biotinylated lentivirus, CD200ED-coreSA-tagged lentivirus, CD200-blocked virus, or left untreated (control). Following incubation, the culture supernatants were collected and analyzed for cytokine levels using commercial enzyme-linked immunosorbent assay (ELISA) kits. Briefly, 96-well microtiter plates were coated with anti-IL-6 or anti-TNF-α capture antibodies and incubated overnight at 4 °C. The plates were then washed three times with wash buffer (PBS containing 0.05% Tween-20) to remove unbound antibodies, followed by blocking with ELISPOT diluent for 1 hour at room temperature. After an additional three washes, standard cytokine solutions (IL-6 and TNF-α) and experimental samples (culture supernatants) were added and incubated for 2 hours at room temperature. The plates were washed again and incubated with detection antibodies (anti-IL-6 or anti-TNF-α) at manufacturer-recommended dilutions for 1 hour at room temperature. After washing, streptavidin-HRP conjugate was added and incubated for 30 minutes at room temperature. Subsequently, tetramethylbenzidine substrate was added to each well, and the reaction was allowed to proceed for 15 minutes in the dark at room temperature. The reaction was terminated by adding 1 M phosphoric acid, and absorbance was measured at 450 nm using a microplate reader (SpectraMax M2e). Cytokine concentrations were determined by interpolation from standard curves generated using the known concentrations of IL-6 and TNF-α.Phagocytosis of biotinylated lentivirus tagged with CD200ED-coreSA
To assess the impact of CD200ED on macrophage activation and viral uptake, J774A.1 macrophages were incubated with biotinylated lentiviral vectors either conjugated with or without CD200ED-coreSA protein. The cells were exposed to the virus for 3 hours, after which the culture medium was replaced. Seventy-two hours post-infection, macrophage cells expressing GFP were analyzed by fluorescence microscopy and flow cytometry to evaluate lentiviral infection rates and phagocytic efficiency. HEK 293T cells, a non-phagocytic cell line, were included as a negative control and subjected to identical treatment and analysis conditions. Additional control experiments included macrophages treated with: (i) biotinylated lentivirus mixed with coreSA protein (lacking CD200ED), and (ii) non-biotinylated lentivirus mixed with CD200ED-coreSA protein. These controls were used to distinguish the specific role of biotin–streptavidin–mediated CD200ED conjugation in modulating phagocytosis by J774A.1 macrophages, compared to CD200ED-coreSA-coated biotinylated lentivirus.CD200/CD200R inhibition assay
To confirm that macrophage downregulation was mediated specifically by the CD200–CD200R interaction, J774A.1 macrophages were treated with CD200ED-modified lentiviral particles under three blocking conditions: (1) viral particles pre-incubated with 10 µg mL−1 anti-mouse CD200 neutralizing antibody (Clone OX-90, BioLegend) to block ligand-receptor binding, (2) macrophages pre-treated with 10 µg mL−1 anti-CD200R antibody (Clone OX-110, BioLegend) for 1 hour at 37 °C to saturate surface receptors, or (3) combined blockade using both antibodies. A control group received unblocked CD200ED-modified lentivirus. Macrophages were seeded in 6-well plates (5 × 105 cells per well) and exposed to viral particles (MOI = 10) for 24 hours. This approach allowed evaluation of the specific role of the CD200–CD200R interaction in mediating the immunomodulatory effects of the engineered viral particles.Statistical analysis
Statistical analyses were performed using GraphPad Prism 9.0. All data, derived from at least three independent biological replicates, are expressed as mean ± standard deviation (SD). Group comparisons were assessed via one-way analysis of variance (ANOVA) followed by Tukey's post hoc test for multiple comparisons. A p value < 0.05 was considered statistically significant.Results
Construction of CD200ED-coreSA encoding plasmid
The gene sequences encoding the CD200 ectodomain (291 bp) and core streptavidin (coreSA, 384 bp) were amplified by PCR and verified by DNA gel electrophoresis. As shown in Fig. 1, the PCR products corresponded to approximately 0.3 kb for CD200ED and 0.4 kb for coreSA. The plasmid construct pME007 was engineered to express the coreSA gene (∼14.2 kDa), cloned between the XhoI and EcoRI restriction sites, and the CD200ED gene (∼10.77 kDa), inserted between the BamHI and EcoRV sites. A short sequence of approximately 6 base pairs (∼0.22 kDa) remained between the EcoRI and BamHI sites (Fig. 1A). This configuration resulted in the expression of the recombinant CD200ED-coreSA fusion protein with an estimated molecular weight of ∼25 kDa.Characterization of recombinant CD200ED-coreSA fusion protein
The plasmid construct encoding the CD200ED-coreSA fusion protein (pME007) was successfully transformed into E. coli Lemo21(DE3) cells and selected using kanamycin-containing agar plates. Following protein expression and purification, elution fractions were collected and monitored by absorbance at 280 nm to generate an elution profile. Most of the recombinant protein was recovered in fraction 9 (Fig. 2A). Analysis of both crude lysate and purified protein by 12% SDS-PAGE, followed by Coomassie staining, revealed a prominent band at ∼25 kDa in the lysate (Fig. 2B), corresponding to the expected molecular weight of the CD200ED-coreSA fusion protein. Elution fractions were concentrated using a centrifugal filter device with a 10 kDa molecular weight cut-off, and protein concentration was determined by BCA assay to be 461 ± 22 µg mL−1. Western blot analysis further confirmed the identity of the recombinant protein. Using anti-His tag, anti-streptavidin, and anti-mouse CD200 antibodies, all three blots showed a single band at ∼25 kDa, verifying the presence and integrity of the CD200ED-coreSA fusion protein (Fig. 2C). It is important to note that coreSA forms a tetramer at 37 °C, resulting in an approximately 100 kDa CD200ED–coreSA fusion protein when tethered to biotinylated viral particles in this study. In contrast, the gel images in Fig. 2 show a single band at ∼25 kDa, corresponding to the monomeric CD200ED–coreSA fusion protein, because the sample was analyzed under denaturing conditions (heated to 95 °C for 5 min). When the fusion protein was incubated at 60 °C, the gel showed four bands (tetramer, trimer, dimer, and monomer; data not shown). If coupling of the CD200ED–coreSA fusion protein to biotinylated viral particles were performed at 60 °C, size-exclusion chromatography would be necessary to isolate a monodisperse protein population. However, because the conjugation carried out at 4 °C was purely tetrameric, this concern is eliminated.Characterization of biotinylated lentivirus tethered with CD200ED-coreSA recombinant protein
The characterization of biotinylated lentivirus tethered with the CD200ED-coreSA recombinant protein was performed using streptavidin-conjugated gold nanoparticles (SA-AuNPs) and transmission electron microscopy (TEM). The biotin-labeled lentivirus (with size about 100 nm) was successfully capped with 10-nm AuNPs, which were observed to bind extensively across the entire viral surface (Fig. 3A). In contrast, the native lentivirus showed no attachment of SA-AuNPs, as expected due to the absence of biotin (Fig. 3B). This confirms the successful biotinylation of the lentivirus and its ability to interact specifically with the purified CD200ED-coreSA fusion protein for the formation of CD200-tagged lentiviral nanoparticles. Dynamic light scattering measurements of size, polydispersity index, and zeta potential of lentivirus are 99.62 ± 8.6 nm, 0.241 ± 0.021, and −18.17 ± 1.8 mV, respectively. Similar measurements were obtained for biotinylated lentivirus (100.02 ± 7.2 nm, 0.252 ± 0.032, and −18.47 ± 2.1 mV) and CD200ED-modifed lentivirus (103.02 ± 10.2 nm, 0.267 ± 0.028, −18.72 ± 1.9 mV). The viral sizes, with both native and modified forms determined by dynamic light scattering, are in line with the scale obtained by TEM images (i.e., ∼100 nm). The zeta potentials were determined to be negative in all types of viral nanoparticles.As shown in Fig. 3A, the Au nanoparticles exhibit a fairly uniform, non-overlapping distribution on the virion surface. Quantitative analysis of five TEM images yielded an average surface coverage of 22 ± 4 AuNPs per two-dimensional projection. Given the absence of nanoparticle overlap, we assume a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry between a CD200ED-coreSA molecule and a biotin anchor on the viral envelope. Since TEM provides only a two-dimensional projection of a spherical virion, we estimate the total number of CD200ED-coreSA molecules per virion to be approximately 88 (i.e., 22 × 4).
1 binding stoichiometry between a CD200ED-coreSA molecule and a biotin anchor on the viral envelope. Since TEM provides only a two-dimensional projection of a spherical virion, we estimate the total number of CD200ED-coreSA molecules per virion to be approximately 88 (i.e., 22 × 4).
Dot blot analysis was conducted to demonstrate the immobilization of CD200ED-coreSA on the surface of biotinylated viral vectors using an anti-mouse CD200 antibody (Fig. 4). The results showed successful antibody binding to both the soluble protein (positive control) and the virus tethered with CD200ED-coreSA. In contrast, no antibody binding was detected for the biotinylated virus alone (negative control), confirming that CD200ED-coreSA had been successfully attached to the surface of the biotinylated viral vectors.
The effect of CD200ED-coreSA on cytokines secretion from J774A.1 macrophage
The effect of CD200ED-coreSA on cytokine secretion from J774A.1 macrophages was evaluated by assessing the levels of two pro-inflammatory cytokines, IL-6 and TNF-α, using ELISA. Macrophages treated with uncoated biotinylated lentivirus secreted 1312 ± 169 pg mL−1 of TNF-α and 52 ± 8 pg mL−1 of IL-6. Similar releasing amounts were detected for cells challenged with coreSA-capped lentivirus, indicating the secretion of IL-6 and TNF-α was not reduced by coreSA moiety or biotin-coreSA affinity binding. In contrast, untreated macrophages (control) secreted significantly less TNF-α (67% reduction) and IL-6 (65% reduction) compared to macrophages infected with uncoated biotinylated lentivirus. When macrophages were treated with CD200ED-coated lentivirus, there was a marked reduction in cytokine secretion: TNF-α secretion decreased by 47.1% to 701 ± 96 pg mL−1, and IL-6 secretion decreased by 55% to 23 ± 3 pg mL−1 (Fig. 5A and B). This reduction in cytokine secretion suggests that CD200ED-coated lentivirus suppresses macrophage activation and exhibits antiphagocytic resistance. However, when the CD200 protein coated on the lentivirus was blocked with anti-CD200 antibody, the activity of the protein was inhibited, resulting in similar cytokine secretion levels as observed in the group treated with biotinylated or coreSA-capped lentivirus. Specifically, the CD200-coated virus blocked with anti-CD200 antibody secreted 1247 ± 179 pg mL−1 of TNF-α and 49 ± 9 pg mL−1 of IL-6.Antiphagocytic efficacy of CD200ED-coreSA tagged on biotinylated lentiviral surface
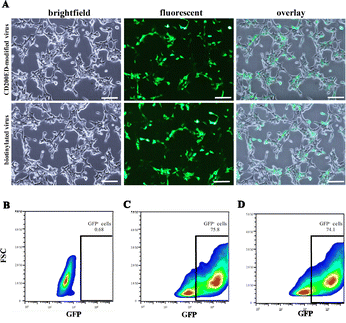
The antiphagocytic efficacy of CD200ED-coreSA tagged on the surface of biotinylated lentiviral particles was evaluated through experiments assessing their effect on macrophage activation. Qualitative results, determined by GFP expression as a measure of viral infectivity, showed that GFP expression was lower in J774A.1 macrophages compared to HEK 293T cells (Fig. 6A and 7A). This observation was further supported by density plots analyzing GFP expression, which revealed significant differences among the groups: untreated negative control (0.57%), unmodified biotinylated virus (61.1%), and CD200ED-modified lentivirus (46%) (Fig. 6B–D). These findings indicate that CD200ED immobilized on the viral surface reduced engulfment by macrophages by approximately 25% compared to unmodified biotinylated virus. The results were further validated using flow cytometry histograms, which demonstrated a mean fluorescence intensity (MFI) of 1.37 × 106 ± 6032 for unmodified biotinylated virus and 9.6 × 105 ± 4102 for CD200ED-modified virus, confirming the reduced phagocytic activity of the modified virus.In contrast, HEK 293T cells infected with CD200ED-coreSA-modified virus do not respond to CD200 signaling. As a result, these cells exhibit greater uptake of viral particles and show similar levels of GFP expression compared to those infected with unmodified biotinylated virus (Fig. 7C and D). This observation is further supported by flow cytometry histograms, which revealed a mean fluorescence intensity (MFI) of 1.94 × 106 ± 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 918 for unmodified biotinylated virus and 1.91 × 106 ± 12
918 for unmodified biotinylated virus and 1.91 × 106 ± 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 for CD200ED-modified virus. However, the reduced phagocytic clearance of CD200ED-modified virus in J774A.1 macrophages is attributed to the interaction of CD200ED with CD200R on the surface of these macrophages. This interaction downregulates macrophage activation and prevents viral entry, thereby enhancing the antiphagocytic efficacy of the modified virus.
106 for CD200ED-modified virus. However, the reduced phagocytic clearance of CD200ED-modified virus in J774A.1 macrophages is attributed to the interaction of CD200ED with CD200R on the surface of these macrophages. This interaction downregulates macrophage activation and prevents viral entry, thereby enhancing the antiphagocytic efficacy of the modified virus.
J774A.1 macrophages were separately infected with coreSA-modified virus and non-biotinylated lentiviral vectors mixed with CD200ED-coreSA protein to further evaluate the interaction of the CD200ED-CD200R axis. It was observed that these two groups could not mediate phagocytosis, likely due to the absence of CD200ED protein or the inability of the CD200ED-coreSA protein to immobilize on the surface of non-biotinylated virus (Fig. 8A). Additionally, the fluorescence intensity of GFP expression in J774A.1 macrophages showed that these two groups exhibited a 3-4-fold higher signal compared to the CD200ED-modified virus group (Fig. 8B). Based on these findings, it is evident that the CD200 ectodomain plays a crucial role in downregulating macrophage activity and inhibiting phagocytosis.
Inhibition of phagocytosis by blocking of CD200–CD200R interaction
The inhibition of phagocytosis was investigated by blocking the interaction between CD200 and its receptor, CD200R. Fig. 9A shows qualitative results indicating that when CD200ED protein immobilized on the lentivirus membrane is blocked by anti-CD200 antibody (ii), when the CD200 receptor located on the macrophage surface is blocked by anti-CD200R antibody (iii), or when both CD200ED on lentivirus and CD200R on macrophages are blocked (iv), there is a significantly higher level of GFP expression compared to the unblocked CD200ED-modified virus (control) (iv). These findings suggest that disrupting the CD200–CD200R axis binding leads to increased engulfment of lentivirus by macrophages, resulting in failure to resist phagocytosis. This observation was further confirmed by measuring the GFP intensity per cell, which demonstrated approximately a 2-fold increase in virus uptake by macrophages in all three blocked groups compared to the unblocked CD200-modified lentivirus group (Fig. 9B).Discussion
Viral vectors are widely leveraged for gene delivery due to their high transduction efficiency, yet their clinical application is constrained by innate and adaptive immune responses to viral components (e.g., capsid proteins, envelope glycoproteins).51 These immune reactions, including neutralizing antibody production and complement activation, accelerate vector clearance and limit therapeutic efficacy.52 To address this, we engineered lentiviral vectors displaying the immunoglobulin-variable (IgV) ectodomain of CD200, an immunoregulatory transmembrane protein that suppresses myeloid cell activation via interaction with its receptor CD200R. CD200R is expressed on macrophages, dendritic cells, and T cells, and its engagement by CD200 triggers inhibitory signaling pathways that reduce pro-inflammatory cytokine release and phagocytic activity.53 This immunosuppressive axis is evolutionarily conserved: CD200 homologs encoded by herpesviruses and poxviruses enable immune evasion by mimicking host CD200.47,48 In murine models, CD200 deficiency exacerbates macrophage activation and microglial phagocytosis, while CD200-Fc fusion proteins inhibit oligodendrocyte precursor engulfment.40 Our findings extend these observations by demonstrating that lentiviral vectors functionalized with the CD200 IgV domain reduce the phagocytosis of murine J774A.1 macrophages. These results align with our previous report that both nano- and micro-sized particles decorated with the ectodomain of CD200 (containing both the IgV and IgC regions) reduced phagocytosis activity in human THP-1 macrophages.42 This reduction in phagocytosis was found to be associated with a downregulation of TLR4 expression on the surface of macrophages.40,42 The CD200–CD200R interaction is mediated entirely by their N-terminal IgV domain, while the IgC domain is positioned near the membrane and contributes structurally rather than to receptor engagement. Multivalent display of the IgV module, such as on particle or surface scaffolds, can promote receptor clustering and generate downstream effects comparable to those elicited by the full ectodomain. In fact, it is surmised that the immunomodulatory potency of IgV domain might be stronger than the full ectodomain when immobilized on surface because it avoids steric hindrance caused by IgC stalk orientation.While most viruses encoding CD200 homologs contain both the IgV and IgC domains, some viruses, such as poxviruses, express proteins like M141R, which contains only the IgV domain.54 The M141R protein is responsible for downregulating macrophage and T-cell activity by binding to CD200R.48,55,56 Similarly, Kaposi's sarcoma-associated herpesvirus encodes the K14 protein, which shares 44% amino acid homology with the human CD200 IgV domain and inhibits myeloid cell activation via CD200R engagement.57,58 Building on these insights, we engineered lentiviral vectors displaying the murine CD200 IgV domain (CD200ED) to mimic viral immune evasion tactics. CD200ED-coreSA fusion proteins was produced via bacterial expression and anchored to biotinylated lentiviral surfaces. A key finding was that while macrophages treated with uncoated biotinylated lentivirus expressed approximately two-thirds more GFP compared to those treated with CD200ED-coated lentivirus, while HEK 293T cells treated in the same manner exhibited the same level of GFP expression. This indicated that the CD200R expressed on the surface of macrophages plays an essential role in regulating phagocytosis and activation upon binding of the CD200 protein. Furthermore, two additional control groups were included: one with coreSA protein alone and another with CD200ED-coreSA interacting with non-biotinylated virus. Both controls showed higher GFP expression compared to the virus tagged with the CD200 protein, indicating that neither successfully prevented phagocytosis or escaped clearance by macrophages. This suggests that either the CD200 protein was missing or failed to attach properly to the virus surface. To confirm the antiphagocytic effect mediated by the CD200–CD200R interaction, experiments were conducted by blocking the immobilization of CD200 protein on the virus membrane and the CD200R on the macrophage surface. Blocking either or both components resulted in increased engulfment of the virus by macrophages, further validating the importance of this interaction. The present study demonstrates that the extracellular domain of CD200 protein can suppress macrophage activation when murine J774A.1 macrophages are treated with biotinylated lentivirus tagged with the CD200 IgV domain. Notably, the antiphagocytic effect of CD200ED modification is less potent than that of CD47 modification.20 However, one could tether viral surface with both CD200ED and CD47 molecules to augment phagocytosis resistance for therapeutic gene delivery. This potential is supported by recent findings demonstrate a synergistic effect when both the CD200–CD200R and CD47-SIRPα axes are blocked, leading to significantly enhanced phagocytosis and tumor growth inhibition.59
Furthermore, the present study capitalized on the known fact that pro-inflammatory cytokines (e.g., TNF-α and IL-6) are secreted by macrophages in response to pathogens, including viruses.60–62 Our findings revealed that macrophages treated with lentivirus decorated with the CD200 IgV extracellular domain protein showed significantly reduced secretion of these pro-inflammatory cytokines by J774A.1 macrophages compared to those treated with biotinylated lentivirus or blocked CD200-capped virus groups, which exhibited much higher levels of TNF-α and IL-6. This confirms the ability of CD200 to modulate macrophage activation and suppress inflammatory responses. Our findings align with the effects observed in CD200-coated micro/nanoparticles tethered the ectodomain of CD200 (containing both IgV and IgC domains).42
Conclusions
Our findings highlight the promise of a biomimetic approach leveraging the CD200–CD200R immune checkpoint. Functionalizing viral vectors with CD200 may enhance gene delivery by attenuating immune responses, minimizing macrophage-mediated clearance, and facilitating targeted delivery of therapeutic genes.Author contributions
Conceptualization: C.-A. P.; methodology, investigation, validation, and visualization: E. A., I. P., S. J., and C.-A. P.; supervision: C.-A. P.; writing – original draft: E. A. and I. P.; writing – review and editing: E. A., I. P., S. J., and C.-A. P.Conflicts of interest
The authors declare no conflicts of interest.Data availability
The data presented in this study are available on request from the corresponding author.Note added after first publication
This article replaces the version published on 22 December 2025 in which the flow cytometry plot in Figure 6B and flow cytometry plot and gating in Figure 6C were not visible. Additionally, a middle initial was added for the author Esmael Alyami, and the footnote listing their current affiliation was updated.Acknowledgements
This study was financially supported by University of Idaho.References
- K. Ikeda, T. Ichikawa, H. Wakimoto, J. S. Silver, T. S. Deisboeck, D. Finkelstein, G. R. Harsh, D. N. Louis, R. T. Bartus, F. H. Hochberg and E. A. Chiocca, Oncolytic virus therapy of multiple tumors in the brain requires suppression of innate and elicited antiviral responses, Nat. Med., 1999, 5, 881–887 Search PubMed.
- H. Wakimoto, K. Ikeda, T. Abe, T. Ichikawa, F. H. Hochberg, R. A. Ezekowitz, M. S. Pasternack and E. A. Chiocca, The complement response against an oncolytic virus is species-specific in its activation pathways, Mol. Ther., 2002, 5, 275–282 Search PubMed.
- H. Sakurai, K. Kawabata, F. Sakurai, S. Nakagawa and H. Mizuguchi, Innate immune response induced by gene delivery vectors, Int. J. Pharm., 2008, 354, 9–15 Search PubMed.
- V. Tsai, D. E. Johnson, A. Rahman, S. F. Wen, D. LaFace, J. Philopena, J. Nery, M. Zepeda, D. C. Maneval, G. W. Demers and R. Ralston, Impact of human neutralizing antibodies on antitumor efficacy of an oncolytic adenovirus in a murine model, Clin. Cancer Res., 2004, 10, 7199–7206 Search PubMed.
- G. Marelli, A. Howells, N. R. Lemoine and Y. Wang, Oncolytic viral therapy and the immune system: A double-edged sword against cancer, Front. Immunol., 2018, 9, 866 Search PubMed.
- A. Lemos de Matos, L. S. Franco and G. McFadden, Oncolytic viruses and the immune system: The dynamic duo, Mol. Ther. Methods Clin. Dev., 2020, 17, 349–358 Search PubMed.
- S. Worgall, G. Wolff, E. Falck-Pedersen and R. G. Crystal, Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration, Hum. Gene Ther., 1997, 8, 37–44 Search PubMed.
- X. Ye, M. Jerebtsova and P. E. Ray, Liver bypass significantly increases the transduction efficiency of recombinant adenoviral vectors in the lung, intestine, and kidney, Hum. Gene Ther., 2000, 11, 621–627 Search PubMed.
- E. K. Mader, Y. Maeyama, Y. Lin, G. W. Butler, H. M. Russell, E. Galanis, S. J. Russell, A. B. Dietz and K. W. Peng, Mesenchymal stem cell carriers protect oncolytic measles viruses from antibody neutralization in an orthotopic ovarian cancer therapy model, Clin. Cancer Res., 2009, 15, 7246–7255 Search PubMed.
- J. García-Castro, R. Alemany, M. Cascalló M, J. Martínez-Quintanilla, M. Arriero Mdel, A. Lassaletta, L. Madero and M. Ramírez, Treatment of metastatic neuroblastoma with systemic oncolytic virotherapy delivered by autologous mesenchymal stem cells: an exploratory study, Cancer Gene Ther., 2010, 17, 476–483 Search PubMed.
- M. A. Croyle, S. M. Callahan, A. Auricchio, G. Schumer, K. D. Linse, J. M. Wilson, L. J. Brunner and G. P. Kobinger, PEGylation of a vesicular stomatitis virus G pseudotyped lentivirus vector prevents inactivation in serum, J. Virol., 2004, 78, 912–921 Search PubMed.
- P. Wonganan and M. A. Croyle, PEGylated adenoviruses: From mice to monkeys, Viruses, 2010, 2, 468–502 Search PubMed.
- E. A. Weaver and M. A. Barry, Effects of shielding adenoviral vectors with polyethylene glycol on vector-specific and vaccine-mediated immune responses, Hum. Gene Ther., 2008, 19, 1369–1382 Search PubMed.
- G. T. Kozma, T. Shimizu and T. Ishida, J. Szebeni. Anti-PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals, Adv. Drug Delivery Rev., 2020, 154–155, 163–175 Search PubMed.
- P. A. Oldenborg, A. Zheleznyak, Y. F. Fang, C. F. Lagenaur, H. D. Gresham and F. P. Lindberg, Role of CD47 as a marker of self on red blood cells, Science, 2000, 288, 2051–2054 Search PubMed.
- H. Wang, J. VerHalen, M. L. Madariaga, S. Xiang, S. Wang, P. Lan, P. A. Oldenborg, M. Sykes and Y. G. Yang, Attenuation of phagocytosis of xenogeneic cells by manipulating CD47, Blood, 2006, 109, 836–842 Search PubMed.
- T. Matozaki, Y. Murata, H. Okazawa and H. Ohnishi, Functions and molecular mechanisms of the CD47-SIRPα signalling pathway, Trends Cell Biol., 2009, 19, 72–80 Search PubMed.
- N. G. Sosale, I. I. Ivanovska, R. K. Tsai, J. Swift, J. W. Hsu, C. M. Alvey, P. W. Zoltick and D. E. Discher, “Marker of Self” CD47 on lentiviral vectors decreases macrophage-mediated clearance and increases delivery to SIRPA-expressing lung carcinoma tumors, Mol. Ther. Methods Clin. Dev., 2016, 3, 16080 Search PubMed.
- M. Milani, A. Annoni, F. Moalli, T. Liu, D. Cesana, A. Calabria, S. Bartolaccini, M. Biffi, F. Russo, I. Visigalli, A. Raimondi, S. Patarroyo-White, D. Drager, P. Cristofori, E. Ayuso, E. Montini, R. Peters, M. Iannacone, A. Cantore and L. Naldini, Phagocytosis-shielded lentiviral vectors improve liver gene therapy in nonhuman primates, Sci. Transl. Med., 2019, 11, 7325 Search PubMed.
- E. M. Alyami, A. Tarar and C. A. Peng, Less phagocytosis of viral vectors by tethering with CD47 ectodomain, J. Mater. Chem B, 2021, 10, 64–77 Search PubMed.
- A. N. Barclay, G. J. Wright, G. Brooke and M. H. Brown, CD200 and membrane protein interactions in the control of myeloid cells, Trends Immunol., 2002, 23, 285–290 Search PubMed.
- G. J. Wright, M. Jones, M. J. Puklavec, J. H. Brown and A. N. Barclay, The unusual distribution of the neuronal/lymphoid cell surface C200 (OX2) glycoprotein is conserved in humans, Immunology, 2001, 102, 173–179 Search PubMed.
- S. G. Meuth, O. J. Simon, A. Grimm, N. Melzer, A. M. Herrmann, P. Spitzer, P. Landgraf and H. Wiendl, CNS inflammation and neuronal degeneration is aggravated by impaired CD200-CD200R-mediated macrophage silencing, J. Neuroimmunol., 2008, 194, 62–69 Search PubMed.
- M. D. Rosenblum, E. B. Olasz, K. B. Yancey, J. E. Woodliff, Z. Lazarova, K. A. Gerber and R. L. Truitt, Expression of CD200 on epithelial cells of the murine hair follicle: a role in tissue-specific immune tolerance?, J. Invest. Dermatol., 2004, 123, 880–887 Search PubMed.
- Y. C. Ko, H. F. Chien, Y. F. Jiang-Shieh, C. Y. Chang, M. H. Pai, J. P. Huang, H. M. Chen and C. H. Wu, Endothelial CD200 is heterogeneously distributed, regulated and involved in immune cell-endothelium interactions, J. Anat., 2009, 214, 183–195 Search PubMed.
- D. Hatherley, S. M. Lea, S. Johnson and A. N. Barclay, Structures of CD200/CD200 receptor family and implications for topology, regulation, and evolution, Structure, 2013, 21, 820–832 Search PubMed.
- R. M. Hoek, S. R. Ruuls, C. A. Murphy, G. J. Wright, R. Goddard, S. M. Zurawski, B. Blom, M. E. Homola, W. J. Streit, M. H. Brown, A. N. Barclay and J. D. Sedgwick, Down-regulation of the macrophage lineage through interaction with OX2 (CD200), Science, 2000, 290, 1768–1771 Search PubMed.
- G. J. Wright, M. J. Puklavec, A. C. Willis, R. M. Hoek, J. D. Sedgwick, M. H. Brown and A. N. Barclay, Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function, Immunity, 2000, 13, 233–242 Search PubMed.
- G. J. Wright, H. Cherwinski, M. Foster-Cuevas, G. Brooke, M. J. Puklavec, M. Bigler, Y. Song, M. Jenmalm, D. Gorman, T. McClanahan, M. R. Liu, M. H. Brown, J. D. Sedgwick, J. H. Phillips and A. N. Barclay, Characterization of the CD200 receptor family in mice and humans and their interactions with CD200, J. Immunol., 2003, 171, 3034–3046 Search PubMed.
- B. T. Kawasaki and W. L. Farrar, Cancer stem cells, CD200 and immunoevasion, Trends Immunol., 2008, 29, 464–468 Search PubMed.
- Y. Liu, Y. Bando, D. Vargas-Lowy, W. Elyaman, S. J. Khoury, T. Huang, K. Reif and T. Chitnis, CD200R1 agonist attenuates mechanisms of chronic disease in a murine model of multiple sclerosis, J. Neurosci., 2010, 30, 2025–2038 Search PubMed.
- L. F. Lue, Y. M. Kuo, T. Beach and D. G. Walker, Microglia activation and anti-inflammatory regulation in Alzheimer's disease, Mol. Neurobiol., 2010, 41, 115–128 Search PubMed.
- G. Manich, M. Recasens, T. Valente, B. Almolda, B. Gonzalez and B. Castellano, Role of the CD200-CD200R axis during homeostasis and neuroinflammation, Neuroscience, 2019, 405, 118–136 Search PubMed.
- C. A. Vaine and R. J. Soberman, The CD200-CD200R1 inhibitory signaling pathway: immune regulation and host-pathogen interactions, Adv. Immunol., 2014, 121, 191–211 Search PubMed.
- K. Kotwica-Mojzych, B. Jodłowska-Jędrych and M. Mojzych, CD200:CD200R interactions and their importance in immunoregulation, Int. J. Mol. Sci., 2021, 22, 1602 Search PubMed.
- A. M. R. van den Bosch, D. Wever, P. Schonewille, S. L. Schuller, J. Smolders, J. Hamann and I. Huitinga, Cortical CD200-CD200R and CD47-SIRPα expression is associated with multiple sclerosis pathology, Brain Commun., 2024, 6, fcae264 Search PubMed.
- R. Feuer, Tickling the CD200 receptor: A remedy for those irritating macrophages, Am. J. Pathol., 2007, 171, 396–398 Search PubMed.
- D. G. Walker, J. E. Dalsing-Hernandez, N. A. Campbell and L. F. Lue, Decreased expression of CD200 and CD200 receptor in Alzheimer's disease: a potential mechanism leading to chronic inflammation, Exp. Neurol., 2009, 215, 5–19 Search PubMed.
- C. Ngwa and F. Liu, CD200-CD200R signaling and diseases: a potential therapeutic target?, Int. J. Physiol. Pathophysiol. Pharmacol., 2019, 11, 297–309 Search PubMed.
- K. Hayakawa, L. D. Pham, J. H. Seo, N. Miyamoto, T. Maki, Y. Terasaki, S. Sakadžić, D. Boas, K. van Leyen, C. Waeber, K. W. Kim, K. Arai and E. H. Lo, CD200 restrains macrophage attack on oligodendrocyte precursors via toll-like receptor 4 downregulation, J. Cereb. Blood Flow Metab., 2016, 36, 781–793 Search PubMed.
- A. Lyons, A. M. Minogue, R. S. Jones, O. Fitzpatrick, J. Noonan, V. A. Campbell and M. A. Lynch, Analysis of the impact of CD200 on phagocytosis, Mol. Neurobiol., 2017, 54, 5730–5739 Search PubMed.
- J. Zhang and C. A. Peng, Diminution of phagocytosed micro/nanoparticles by tethering with immunoregulatory CD200 protein, Sci. Rep., 2020, 10, 1–13 Search PubMed.
- R. Ocaña-Guzman, L. Vázquez-Bolaños and I. Sada-Ovalle, Receptors that inhibit macrophage activation: Mechanisms and signals of regulation and tolerance, J. Immunol. Res., 2018, 2018, 8695157 Search PubMed.
- R. Mihrshahi, A. N. Barclay and M. H. Brown, Essential roles for Dok2 and RasGAP in CD200 receptor-mediated regulation of human myeloid cells, J. Immunol., 2009, 183, 4879–4886 Search PubMed.
- S. Zhang, H. Cherwinski, J. D. Sedgwick and J. H. Phillips, Molecular mechanisms of CD200 inhibition of mast cell activation, J. Immunol., 2004, 173, 6786–6793 Search PubMed.
- H. Shinohara, A. Inoue, N. Toyama-Sorimachi, Y. Nagai, T. Yasuda, H. Suzuki, R. Horai, Y. Iwakura, T. Yamamoto, H. Karasuyama, K. Miyake and Y. Yamanashi, Dok-1 and Dok-2 are negative regulators of lipopolysaccharide-induced signaling, J. Exp. Med., 2005, 201, 333–339 Search PubMed.
- I. Shiratori, M. Yamaguchi, M. Suzukawa, K. Yamamoto, L. L. Lanier, T. Saito and H. Arase, Down-regulation of basophil function by human CD200 and human herpesvirus-8 CD200, J. Immunol., 2005, 175, 4441–4449 Search PubMed.
- C. M. Cameron, J. W. Barrett, L. Liu, A. R. Lucas and G. McFadden, Myxoma virus M141R expresses a viral CD200 (vOX-2) that is responsible for down-regulation of macrophage and T-cell activation in vivo, J. Virol., 2005, 79, 6052–6067 Search PubMed.
- S. Dübel, F. Breitling, R. Kontermann, T. Schmidt, A. Skerra and M. Little, Bifunctional and multimeric complexes of streptavidin fused to single chain antibodies (scFv), J. Immunol. Methods, 1995, 178, 201–209 Search PubMed.
- A. Tarar, E. M. Alyami and C. A. Peng, Efficient expression of soluble recombinant protein fused with core-streptavidin in bacterial strain with T7 expression system, Methods Protoc., 2020, 3, 82 Search PubMed.
- A. D. Miller, Retrovirus packaging cells, Hum. Gene Ther., 1990, 1, 5–14 Search PubMed.
- P. U. Emeagi, C. Goyvaerts, S. Maenhout, J. Pen, K. Thielemans and K. Breckpot, Lentiviral vectors: A versatile tool to fight cancer, Curr. Mol. Med., 2013, 13, 602–625 Search PubMed.
- M. C. Jenmalm, H. Cherwinski, E. P. Bowman, J. H. Phillips and J. D. Sedgwick, Regulation of myeloid cell function through the CD200 receptor, J. Immunol., 2006, 176, 191–199 Search PubMed.
- D. Farré, P. Martínez-Vicente, P. Engel and A. Angulo, Immunoglobulin superfamily members encoded by viruses and their multiple roles in immune evasion, Eur. J. Immunol., 2017, 47, 780–796 Search PubMed.
- L. Zhang, M. Stanford, J. Liu, C. Barrett, L. Jiang, A. N. Barclay and G. McFadden, Inhibition of macrophage activation by the myxoma virus M141 protein (vCD200), J. Virol., 2009, 83, 9602–9607 Search PubMed.
- M. Akkaya, L. S. Kwong, E. Akkaya, D. Hatherley and A. N. Barclay, Rabbit CD200R binds host CD200 but not CD200-like proteins from poxviruses, Virology, 2016, 488, 1–8 Search PubMed.
- M. Foster-Cuevas, G. J. Wright, M. J. Puklavec, M. H. Brown and A. N. Barclay, Human herpesvirus 8 K14 protein mimics CD200 in down-regulating macrophage activation through CD200 receptor, J. Virol., 2004, 78, 7667–7676 Search PubMed.
- S. A. Rezaee, J. A. Gracie, I. B. McInnes and D. J. Blackbourn, Inhibition of neutrophil function by the Kaposi's sarcoma-associated herpesvirus vOX2 protein, AIDS, 2005, 19, 1907–1910 Search PubMed.
- J. Li, Z. Wang, X. Qin, M. C. Zhong, Z. Tang, J. Qian, J. Dou, T. Hussell, P. D. King, J. A. Nunès, Y. Yamanashi, D. Davidson and A. Veillette, CD200R1-CD200 checkpoint inhibits phagocytosis differently from SIRPα-CD47 to suppress tumor growth, Nat. Commun., 2025, 16, 5145 Search PubMed.
- R. Z. Murray, J. G. Kay, D. G. Sangermani and J. L. Stow, A role for the phagosome in cytokine secretion, Science, 2005, 310, 1492–1495 Search PubMed.
- A. Aderem, Phagocytosis and the inflammatory response, J. Infect. Dis., 2003, 187, S340–S345 Search PubMed.
- A. P. Manderson, J. G. Kay, L. A. Hammond, D. L. Brown and J. L. Stow, Subcompartments of the macrophage recycling endosome direct the differential secretion of IL-6 and TNFα, J. Cell Biol., 2007, 178, 57–69 Search PubMed.
Footnote |
| † Currently at Department of Biology, King Khalid University, P. O. Box 9004, Abha 61413, Saudi Arabia. |
| This journal is © The Royal Society of Chemistry 2026 |