Open Access Article
Open Access ArticleAn integrated approach of green chemistry and analytical quality-by-design for simultaneous estimation of drugs in a fixed-dose combination by a stability-indicating RP-HPLC method
Varsha Vishnupant
Thorat
 a,
Sudhakar Maruti
Alave
a,
Sudhakar Maruti
Alave
 *a and
Vijay Arjun
Bagul
*a and
Vijay Arjun
Bagul
 b
b
aMaharshi Dayanand College of Arts, Science and Commerce, Mumbai-400012, Maharashtra, India. E-mail: sudhakaralave@gmail.com
bThe Institute of Science, Dr Homi Bhabha State University, Madam Cama Road, Mumbai-400032, Maharashtra, India
First published on 2nd January 2026
Abstract
The RP-HPLC analytical method has been developed and validated for the simultaneous quantification of chlorthalidone (CHL), amlodipine (AML) and telmisartan (TEL) in a combination drug formulation. An analytical quality-by-design (AQbD) framework was applied using three critical method variables—pump flow rate, oven temperature and pH—to systematically evaluate their influence on chromatographic performance. The mobile phase consists of a water and ethanol mixture and is free from any toxic solvents. Response surface and contour plots enabled robust optimization with final conditions of ethanol–water (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 v/v) containing 0.1% triethylamine and adjusted to pH 2.3 with orthophosphoric acid. Chromatographic separation was achieved on a Sunniest C18 column (150 mm, 4.6 mm, 5 µm) under isocratic conditions. The method operated at a column flow rate of 1.6 mL min−1 with a photodiode array detector. The method was performed using an AQbD-driven response surface central composite design (CCD). The validated method exhibited excellent linearity within 50–150% of the target concentration with correlation coefficients (r) > 0.99 for all analytes. Accuracy studies showed overall mean recoveries of 101.4% for CHL, 100.1% for TEL and 100.4% for AML. Stress degradation under acidic, basic, oxidative, thermal, and photolytic conditions demonstrated that the method is stability-indicating with complete resolution of degraded products and confirmed spectral purity for all analytes. The ecological impact of the new analytical method was evaluated using a variety of greenness assessment techniques. Scores obtained were: AGREE (0.75), MoGAPI (79), BAGI (77.5), AGREE-Prep (0.78), RAPI (55.0), and AMGS-tool (80.96). These figures demonstrate a procedure that complies with green analytical principles with respect to solvent safety, low toxicity and decreased environmental impact. The moderate RAPI score highlighted opportunities for improvement related to energy consumption and instrument efficiency. The proposed RP-HPLC method is precise, accurate, and stability-indicating, and also environmentally responsible, offering a robust and sustainable analytical approach for the simultaneous estimation of drugs in fixed-dose formulations.
65 v/v) containing 0.1% triethylamine and adjusted to pH 2.3 with orthophosphoric acid. Chromatographic separation was achieved on a Sunniest C18 column (150 mm, 4.6 mm, 5 µm) under isocratic conditions. The method operated at a column flow rate of 1.6 mL min−1 with a photodiode array detector. The method was performed using an AQbD-driven response surface central composite design (CCD). The validated method exhibited excellent linearity within 50–150% of the target concentration with correlation coefficients (r) > 0.99 for all analytes. Accuracy studies showed overall mean recoveries of 101.4% for CHL, 100.1% for TEL and 100.4% for AML. Stress degradation under acidic, basic, oxidative, thermal, and photolytic conditions demonstrated that the method is stability-indicating with complete resolution of degraded products and confirmed spectral purity for all analytes. The ecological impact of the new analytical method was evaluated using a variety of greenness assessment techniques. Scores obtained were: AGREE (0.75), MoGAPI (79), BAGI (77.5), AGREE-Prep (0.78), RAPI (55.0), and AMGS-tool (80.96). These figures demonstrate a procedure that complies with green analytical principles with respect to solvent safety, low toxicity and decreased environmental impact. The moderate RAPI score highlighted opportunities for improvement related to energy consumption and instrument efficiency. The proposed RP-HPLC method is precise, accurate, and stability-indicating, and also environmentally responsible, offering a robust and sustainable analytical approach for the simultaneous estimation of drugs in fixed-dose formulations.
Sustainability spotlightThis work presents an environmentally responsible RP-HPLC method developed within an analytical quality by design (AQbD) framework for the simultaneous quantification of chlorthalidone, amlodipine, and telmisartan in combination drug formulations. The method employs an ethanol–water mobile phase system completely free from toxic organic solvents, significantly minimizing environmental hazards and operator exposure. Comprehensive greenness assessments (AGREE = 0.75, MoGAPI = 79, BAGI = 77.5, AGREE-Prep = 0.78, RAPI = 55.0, and AMGS-tool = 80.96) confirm the method's strong alignment with green analytical chemistry principles, demonstrating low solvent toxicity, reduced waste generation, and minimal ecological impact. This study exemplifies how sustainable solvent selection, statistical design optimization, and resource-efficient analytical practices can be integrated to deliver a robust, precise, and stability-indicating method that advances greener pharmaceutical quality control. |
Introduction
Amlodipine besylate, telmisartan and chlorthalidone are combined in a single fixed-dose formulation to provide a synergistic approach to the long-term management of hypertension, particularly in patients requiring multi-mechanistic intervention. The combination unites three pharmacological classes: amlodipine as a dihydropyridine calcium channel antagonist, telmisartan as an angiotensin II receptor (AT1) blocker, and chlorthalidone as a thiazide-related diuretic. This multi-target therapy facilitates more effective and sustained blood pressure control, while also offering additional cardiovascular protection.1Amlodipine besylate is chemically designated as 3-ethyl 5-methyl (±)-2-[(2-aminoethoxy) methyl]-4-(2-chlorophenyl)-6-methyl-1,4-dihydropyridine-3,5-dicarboxylate benzene sulfonate.2 Telmisartan is chemically described as 4′-[(1,4′-dimethyl-2′-propyl[2,6′-bi-1H-benzimidazol]-1′-yl)methyl]-[1,1′-biphenyl]-2-carboxylic acid.3 Chlorthalidone, chemically known as 2-chloro-5-(1-hydroxy-3-oxo isoindolinyl)benzenesulfonamide, is a white crystalline powder with low aqueous solubility, but is soluble in methanol.4
Structural representations of amlodipine besylate, telmisartan, and chlorthalidone are shown in Fig. 1.
High-performance liquid chromatography (HPLC) is the technique most frequently employed for simultaneous estimation of drugs in fixed-dose combinations due to its superior resolution, reproducibility, and sensitivity.5–7 Acetonitrile and methanol are commonly used organic solvents in HPLC mobile phase preparation, though they negatively affect the health of analysts and are harmful to the environment. Compared with conventional HPLC solvents, ethanol provides a balanced chromatographic and environmental profile. Acetonitrile offers the highest elution strength and lowest viscosity, enabling faster separations, but it is petrochemical-derived and associated with greater environmental and toxicological concerns. Methanol provides moderate elution strength with slightly higher viscosity than acetonitrile, yet it is still more toxic and less sustainable than ethanol. Despite having slightly lower elution strength and a higher viscosity, ethanol is far safer, less poisonous, biodegradable and renewable, which improves laboratory safety and decreases its impact on the environment.
In the present study, the UV cut-off for ethanol (∼210 nm) is not an issue as the measurements are carried out at 245 nm.8
Normally, building an analytical technique by HPLC involves a quality by testing (QbT) approach, which includes the changing one factor at a time (OFAT) method. The usual method involves changing parameters during trial-and-error until success is reached. This leads to many experiments, and the collection of the knowledge gained may offer limited insight and be non-reproducible. It can also waste considerable time and resources. Moreover, pharmaceutical companies have been worried about unreliable techniques for years. AQbD provides a methodical approach to address these concerns, focusing on the understanding of the relationship between critical process parameters (CPPs) and critical quality attributes (CQAs) in method development. This innovative method integrates quality-by-design (QbD) principles into the design and scoping of analytical processes to ensure the quality and robustness of the analytical techniques employed.9 In response to this, the ICH has been formulating guidelines. ICH Q14 encourages the replacement of traditional practices with analytical quality by design (AQbD) and provides a risk-based approach to the development of analytical processes.10–13
The most important advantages derived from AQbD approaches depend on the knowledge amassed during the development phase and defining the method operable design region (MODR) to enable validating and selecting a specific operating point for quantitative analysis. Numerous studies on AQbD have demonstrated its advantages in analytical method development.14,15
AQbD focuses on incorporating quality throughout the method development process rather than relying exclusively on post-development validation. In the AQbD cycle, the first step is to set the analytical target profile (ATP). Subsequently, critical method parameters (CMPs) that can affect critical quality attributes (CQAs) such as resolution, retention time, peak shape and method robustness are identified. CMPs include parameters, such as buffer selection, the pH of the aqueous phase, the type and amount of organic modifier, flow rate and temperature of the column. The influence of these parameters on CQAs is then assessed and an appropriate method-design space is developed using systematic experimental designs such as central composites, full factorials, fractional factorials and Box–Behnken designs.
Once the method is developed and validated, researchers direct their attention towards green analytical chemistry (GAC) as a means to protect the environment.16 The primary objective of GAC is to shield both individuals and the environment from harmful waste. Analytical chemists play an active role in promoting the principles of sustainable development through the implementation of GAC. The green analytical procedure index (GAPI), analytical greenness metric (AGREE), metric for evaluating the “greenness” of analytical sample preparation (AGREE-Prep), analytical method greenness score calculator (AMGS), red analytical performance index (RAPI) and blue applicability grade index (BAGI) are the most applied green assessment tools.17–23
A variety of HPLC and LC-MS techniques for amlodipine, telmisartan and chlorthalidone combinations are described in literature studies, which include the use of organic solvents such as acetonitrile and methanol, and some techniques impede routine usage by requiring long, complicated gradients or phosphate buffers for separation.24–39 A comparison of the methodologies used in the literature review is provided in Table 1.
| Sr. no. | Instrument | Molecule combination(s) | Mobile phase (MP) | Run time | Resolution | Reference no. |
|---|---|---|---|---|---|---|
| a TEL = telmisartan, AML = amlodipine, CHT = chlorthalidone, HCTZ = hydrochlorothiazide, ROS = rosuvastatin, ATV = atorvastatin, OLM = olmesartan, ACN = acetonitrile, MeOH = methanol. | ||||||
| 1 | RP-HPLC | TEL + ROS + AML | Methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ACN ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (pH 3.5) water (pH 3.5) |
Run time = 20 min | Resolution > 5 | 24 |
| 2 | RP-HPLC | HCT + AML + TEL | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetate buffer pH 5 (60 acetate buffer pH 5 (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) 40) |
Run time = about 15 min | Resolution > 3.0 | 25 |
| 3 | RP-HPLC | HCT + AML + TEL | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetate buffer pH 5 (60 acetate buffer pH 5 (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) 40) |
Run time = 14 min | Resolution > 2.0 | 26 |
| 4 | RP-HPLC (gradient) | Azelnidipine + telmisartan | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer (25 buffer (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75) 75) |
Run time = 12 min | Resolution > 3.0 | 27 |
| 5 | RP-HPLC | TEL + AML + HCT | ACN/phosphate buffer pH 3.0 | Run time = 12 min | Resolution > 1.5 | 28 |
| 6 | RP-HPLC (AQbD) | TEL + CHT + AML + ATV | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer pH 6.5 (42 buffer pH 6.5 (42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 58) 58) |
Run time = 10 min | Resolution > 1.5 | 29 |
| 7 | RP-HPLC | TEL + CHT + AML | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 mM buffer pH 3.0 (35 20 mM buffer pH 3.0 (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65) 65) |
Run time = about 10 min | Resolution > 3.5 | 30 |
| 8 | RP-HPLC | HCT + AML + TEL | KH2PO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ACN ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (45 MeOH (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) 10) |
Run time = 10 min | Resolution > 2.0 | 31 |
| 9 | RP-HPLC | Efonidipine (EHE) + TEL | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 mM buffer pH 4.9 (45 25 mM buffer pH 4.9 (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55) 55) |
Run time = about 9 min | Resolution > 5.0 | 32 |
| 10 | RP-HPLC | TEL + ATV | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer pH 3.5 (60 buffer pH 3.5 (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) 40) |
Run time = about 8 min | Resolution > 5.0 | 33 |
| 11 | RP-HPLC | AML + CHT + OLM | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer pH 3.0 (60 buffer pH 3.0 (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) 40) |
Run time = about 7 min | Resolution > 3.0 | 34 |
| 12 | RP-HPLC (AZL + TEL) | Azelnidipine + TEL | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 mM phosphate buffer pH 4.6 (70 5 mM phosphate buffer pH 4.6 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) 30) |
Run time = 7 min | Resolution > 3.0 | 35 |
| 13 | RP-HPLC | TEL + AML | Buffer pH 3.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ACN (60 ACN (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) 40) |
Run time = about 5 min | Resolution > 1.5 | 36 |
| 14 | RP-HPLC (binary) | AML + CHT | Gradient water pH 3/ACN | Run time = about 5 min | Resolution > 1.5 | 37 |
| 15 | UPLC | TEL + AML + HCT | Gradient buffer pH 3.2/ACN | Run time = about 5 min | Resolution > 5.0 | 38 |
| 16 | Rapid RP-HPLC | TEL + HCZ + OLM + AML | ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer (45 buffer (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35) 35) |
Run time = about 4.5 min | Resolution > 3.5 | 39 |
In reversed-phase HPLC, the choice of organic modifier plays a crucial role in analyte retention and resolution. Protic alcohols such as ethanol reduce the strength of analyte–stationary phase interactions, leading to shorter retention times, while simultaneously influencing hydrogen bonding due to their dual role as proton donors and acceptors.40 HPLC is also used in the field of quality evaluation and control of natural products.41–45
The present study focuses on the use of AQbD to provide a methodical and scientific approach to the development of a stability-indicating RP-HPLC technique for the simultaneous quantification of telmisartan (TEL), amlodipine (AML) and chlorthalidone (CHL) in a fixed-dose combination. Ethanol was selected as the organic modifier at 35% (v/v) owing to its balanced elution strength, lower viscosity, reduced backpressure, and favourable peak symmetry. As analyte ionization critically governs retention, the mobile phase pH was adjusted to the acidic range, thereby ensuring protonation of basic analytes, suppression of silanol activity and improvement in peak shape. Preliminary univariate trials revealed that pump flow rate, oven temperature and pH were particularly important in governing separation efficiency; hence, these factors were prioritized in subsequent optimization. Forced-degradation studies were performed to demonstrate the specificity and stability-indicating nature of the analytical method, with no co-elution observed between degradation products and analyte peaks.46
The method developed is validated. The ICH Q2(R1)/Q2A guidelines were followed in the validation of the analytical procedure. Specificity, linearity, accuracy, precision (including repeatability and intermediate precision), range, robustness, system appropriateness, and solution/sample stability were all covered in the validation process.
Analytical greenness (AGREE), the multi-objective green analytical procedure index (MoGAPI), BAGI, RAPI, AGREE-Prep, and AMGS-tool were also used to evaluate the method from an environmental sustainability perspective, confirming its compliance with the green chemistry principles of environmentally friendly pharmaceutical analysis. In addition to its analytical advantages, the method incorporates key green chemistry principles through the use of ethanol, reduced solvent consumption, and minimized waste generation. These features directly align with several United Nations Sustainable Development Goals (SDGs), including SDG 3 (Good Health and Well-being) by reducing toxic solvent exposure, SDG 6 (Clean Water and Sanitation) by lowering aquatic toxicity, SDG 12 (Responsible Consumption and Production) through safer and more sustainable solvent use, and SDG 13 (Climate Action) by decreasing the overall carbon and environmental footprint compared with conventional solvents.
To summarise, this study presents the synergistic effect of quality by design (QbD) and green analytical chemistry in the development of a sustainable chromatographic method for the simultaneous determination of chlorthalidone (CHL), amlodipine (AML) and telmisartan (TEL) in a combined drug sample.†
Experimental
Reagents, materials and equipment
Analytical-grade reagents, namely triethylamine, orthophosphoric acid and ethanol were purchased from Rankem chemicals. Samples were received from the Mumbai medical store.Chlorthalidone (CHL), amlodipine (AML) and telmisartan (TEL) standards were purchased from Venus lab. The instrumentation employed was a Thermo-Fisher Ultimate 3000 HPLC system, Lab-india pH meter and analytical weighing balance (Sartorius).
Preparation of the mobile phase
Chromatographic separation was achieved on an Sunniest C18 column (150 mm × 4.6 mm, 5 µm) under isocratic conditions with a green mobile phase consisting of ethanol and water (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65, v/v) containing 0.1% triethylamine, adjusted to pH 2.3 using 1% orthophosphoric acid. The method operated at a flow rate of 1.6 mL min−1 with detection at 245 nm using a photodiode array detector. The mobile phase consisted of ethanol and water (35
65, v/v) containing 0.1% triethylamine, adjusted to pH 2.3 using 1% orthophosphoric acid. The method operated at a flow rate of 1.6 mL min−1 with detection at 245 nm using a photodiode array detector. The mobile phase consisted of ethanol and water (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65, v/v) containing 0.1% triethylamine, adjusted to pH 2.3 using 1% orthophosphoric acid.
65, v/v) containing 0.1% triethylamine, adjusted to pH 2.3 using 1% orthophosphoric acid.
Preparation of the standard solution
Telmisartan stock solution: 40 mg of telmisartan, were weighed and transferred into a 50 mL volumetric flask. 30 mL of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) was added and sonicated to dissolve, then the volume was made up with ethanol
50) was added and sonicated to dissolve, then the volume was made up with ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50). Amlodipine besylate and chlorthalidone stock solution: 63 mg of chlorthalidone and 34 mg of amlodipine besylate were weighed and transferred into a 50 mL volumetric flask. 30 mL of ethanol
50). Amlodipine besylate and chlorthalidone stock solution: 63 mg of chlorthalidone and 34 mg of amlodipine besylate were weighed and transferred into a 50 mL volumetric flask. 30 mL of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) was added and sonicated to dissolve, then the volume was made up with ethanol
50) was added and sonicated to dissolve, then the volume was made up with ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50). Standard solution: 5 mL of telmisartan stock solution and 1 mL of amlodipine besylate and chlorthalidone stock solution were pipetted into a 25 mL volumetric flask, and diluted to volume with the mobile phase.
50). Standard solution: 5 mL of telmisartan stock solution and 1 mL of amlodipine besylate and chlorthalidone stock solution were pipetted into a 25 mL volumetric flask, and diluted to volume with the mobile phase.
Preparation of the sample solution
20 tablets were weighed accurately, and the average weight was calculated. 20 tablets were crushed by a suitable means to a fine powder. Tablet powder equivalent to one tablet was weighed and transferred to a 100 mL volumetric flask, sonicated for 20 min after adding 30 mL of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) with intermittent shaking, diluted with the same solvent to the mark, and mixed well. The sample solution was centrifuged at 4000 rpm for 5 min and 10 mL of supernatant solution was transferred into 25 mL volumetric flask and diluted to volume with mobile phase. The resuting solution was filtered through a 0.45 µm filter discarding the first 2 mL of filtrate.
50) with intermittent shaking, diluted with the same solvent to the mark, and mixed well. The sample solution was centrifuged at 4000 rpm for 5 min and 10 mL of supernatant solution was transferred into 25 mL volumetric flask and diluted to volume with mobile phase. The resuting solution was filtered through a 0.45 µm filter discarding the first 2 mL of filtrate.
The validation parameters described below.
Suitability of system: experimental variables like theoretical plates, asymmetry and percentage rsd of peak area are evaluated from a standard solution.
Linearity and range: for the assessment of linearity, five linearity standards within the range of 50% to 150% of the target concentration were made. A plot of peak area vs. concentration was prepared, each was injected in triplicate, and the regression equation was found. A correlation coefficient (r) indicates acceptable linearity. This value is NLT 0.99.
Accuracy (recovery): a spiked pseudo matrix was used, with known active content percentages at 50%, 100% and 150% of the limit level. The recovery percentage must be established after each stage and is analysed three times. The average recovery is expected to be between 98% and 102%, which are the predetermined acceptable limits.
Robustness: each procedure parameter was consciously changed separately (e.g. column temperature ±2 °C, and flow rate ±0.2 mL min−1). The impact on essential peak parameters was computed.
Filter study: an unfiltered control sample was prepared, and the same solution was filtered through the designated membrane filters (0.45-micron filters). The assay results were compared to check that the filter does not introduce artifacts or adsorb any active constituents.
No significant change in assay values was observed upon reanalysis of the sample and standard solutions at defined time intervals, indicating that the solutions remained constant and stable for the duration of the analysis.
Forced-degradation study: forced-degradation studies confirm specificity by generating degradation products under different stress conditions, to evaluate assay recovery and peak purity. The optimized stress conditions are as follows:
Acid degradation of the sample
Crushed tablets equivalent to the average weight were weighed into a 100 mL volumetric flask. Then about 30 mL of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) was added, and the mixture was shaken and sonicated for 20 min. 5 mL of 0.1 N hydrochloric acid was further added, and the mixture was heated for about an hour at 80 °C. After cooling, it was neutralized with 0.1 N NaOH. The volume was adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well, and then diluted to the mark with the mobile phase.
50) was added, and the mixture was shaken and sonicated for 20 min. 5 mL of 0.1 N hydrochloric acid was further added, and the mixture was heated for about an hour at 80 °C. After cooling, it was neutralized with 0.1 N NaOH. The volume was adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well, and then diluted to the mark with the mobile phase.
Base degradation of the sample
Crushed tablets equivalent to the average weight were weighed into a 100 mL volumetric flask. Then about 30 mL of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) were added, and the mixture was shaken and sonicated for 20 min. 5 mL of 0.1 N NaOH were further added, and the mixture was heated for about an hour at 80 °C. After cooling, it was neutralized with 0.1 N hydrochloric acid. The volume was adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well, and then diluted to the mark with the mobile phase.
50) were added, and the mixture was shaken and sonicated for 20 min. 5 mL of 0.1 N NaOH were further added, and the mixture was heated for about an hour at 80 °C. After cooling, it was neutralized with 0.1 N hydrochloric acid. The volume was adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well, and then diluted to the mark with the mobile phase.
Oxidative degradation of the sample
Crushed tablets equivalent to the average weight were weighed into a 100 mL volumetric flask. Then about 30 mL of the diluent was added, and the mixture was shaken and sonicated for 20 min. 5 mL of 30% H2O2 were further added, and the mixture was heated for about an hour at 80 °C. After cooling, the volume was adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well, and then diluted to the mark with the mobile phase.Photolytic degradation of the sample
For the sample used to assess degradation through photolysis, twenty tablets were crushed and exposed to light at 200 watt-hours per square meter and 1.2 million lux hours.Preparation of the thermal degradation sample
Tablets were subjected to heating at 110 °C for 72 h. Further crushed tablets equivalent to the average weight were weighed into a 100 mL measuring flask, then about 30 mL of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (50
methanol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) was added, and the mixture was shaken and sonicated for 20 min. The volume was further adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well and then diluted to the mark with the mobile phase.
50) was added, and the mixture was shaken and sonicated for 20 min. The volume was further adjusted with diluent and mixed. The sample was centrifuged for 5 min at 4000 rpm. 10 mL of the sample was placed into a 25 mL volumetric flask, mixed well and then diluted to the mark with the mobile phase.
Assessment of greenness by green metric tools
It is necessary for the development of current analytical methods to minimize toxic solvents, chemical waste, price, operation modes and power consumption. At the same time, concerns such as energy usage and cost-effectiveness must also be monitored to ensure overall sustainability.To evaluate the environmental profile of the optimized HPLC method, a comprehensive assessment was performed using multiple green chemistry evaluation tools, each offering a distinct perspective on sustainability. A holistic score of 0.75 was supported by a visual circular diagram that showed strong adherence to most of the 12 principles of green chemistry AGREE, especially concerning the safety of solvents and the minimization of waste. Exploring methods beyond environmental parameters, the MoGAPI index provided a score of 79, demonstrating operational feasibility and a balance of moderate economic concerns mainly around energy spend. In the same light, the BAGI tool also reflected positively on the method, with a score of 77.5, illustrating a good balance between analytical environmental responsibility and operational efficiency. The AGREE-Prep tool (score 0.78) also reflected green sample preparation steps, while the RAPI tool provided a score of 55.0, which was relatively low, indicating a need to improve on energy efficiency and run time efficiency. Finally, low reagent toxicity and high compliance with safer solvent criteria have been confirmed by the AMGS-tool, which reported a remarkable score of 80.96.
Results and discussion
Method development
Initial method development was performed using a mobile phase comprising water and ethanol, aiming to achieve acceptable separation and peak shapes for the analytes. However, early trials revealed a suboptimal peak shape for amlodipine and inadequate separation. To improve both separation and peak symmetry, the aqueous component of the mobile phase was modified by adjusting the pH with orthophosphoric acid (OPA) and incorporating triethylamine (TEA) as a peak shape modifier. The controlled pH of the aqueous phase not only improved the chromatographic behaviour of amlodipine but also contributed to enhanced reproducibility and minimized variability during routine analysis. This strategic modification of the mobile phase resulted in robust and reliable chromatographic performance in subsequent analytical trials. The proposed RP-HPLC method demonstrated overall superior performance compared with previously reports in terms of resolution, run time and overall greenness. The method enabled complete baseline separation of chlorthalidone, amlodipine, and telmisartan, along with their respective degradation products, with a resolution of minimum 3.15 and maximum 6.58, and relatively short runtime with elution of all three peaks within 8 minutes, thereby minimizing solvent usage and reducing environmental impact. A 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ethanol–methanol mixture was selected as the diluent for stock sample preparation because it significantly enhanced the solubility of all analytes, particularly telmisartan, which exhibits limited aqueous solubility, thereby improving the extraction efficiency of the sample. Furthermore, ethanol
50 ethanol–methanol mixture was selected as the diluent for stock sample preparation because it significantly enhanced the solubility of all analytes, particularly telmisartan, which exhibits limited aqueous solubility, thereby improving the extraction efficiency of the sample. Furthermore, ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water as the mobile phase system resulted in sharp peak separation. Thus both solvent systems were found to be compatible and appropriate for the analytes being separated.
water as the mobile phase system resulted in sharp peak separation. Thus both solvent systems were found to be compatible and appropriate for the analytes being separated.
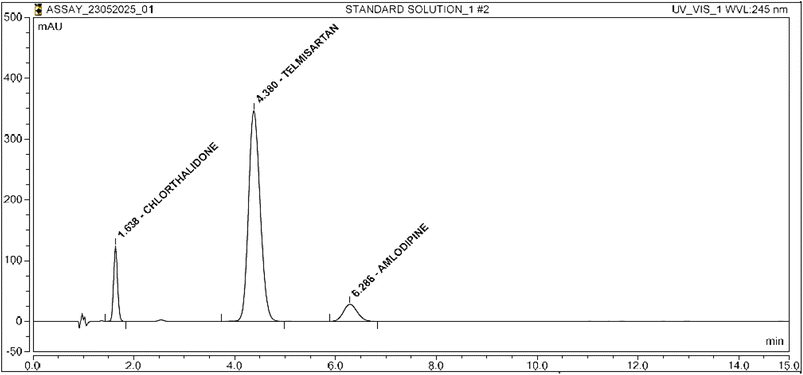
Representative graphs are shown in Fig. 2–5.
Method development and optimization
A quality-by-design (QbD) approach using a central composite design (CCD) was applied to systematically optimize chromatographic conditions for the simultaneous estimation of telmisartan, amlodipine and chlorthalidone. Four critical method attributes—flow rate (1.20–1.70 mL min−1), column temperature (25.0–40.0 °C), pH (2.30–2.50) and detection wavelength—were varied at three levels each, resulting in 17 different experimental runs generated through Design-Expert. The method was developed on a C18 column with dimensions of 4.6 mm × 150 mm under isocratic conditions with 35% ethanol as the organic phase and an injection volume of 20 µL. The analytical target profile was defined in terms of the resolution between critical peak pairs, theoretical plates, and peak asymmetry, all of which served as system suitability parameters.Statistical evaluation confirmed the quadratic model to be significant (p < 0.05), with flow rate and pH emerging as the factors with the most impact on the resolution, while column temperature and wavelength had moderate secondary impact factors. Several sets of operating conditions that balanced resolution, efficiency, and peak symmetry were made available inside the design space via the numerical optimization tool. There were three representative optima. Predicting an Rs-map of 5.310 with cumulative desirability of 0.3651, optimum A (flow 1.583 mL min−1, temperature 25 °C, pH 2.30) was considered first; optimum B (flow 1.200 mL min−1, temperature 37.9 °C, pH 2.30) was best overall for separation, yielding an Rs-map of 6.199, and theoretical plates of 3908 (first peak) and 3871 (last peak) with asymmetries of 1.13 and 1.25 (cumulative desirability of 0.3494); finally, for optimum C (flow 1.200 mL min−1, temperature 37.7 °C, pH 2.50) an Rs-map of 4.916 was derived with plates and symmetry values slightly lower than those for optimum B. Based on the central composite design (CCD), optimum A (flow rate 1.583 mL min−1, column temperature 25 °C, pH 2.30) was the most prioritized and subsequently, finalized condition for routine use, which predicted an Rs-map of 5.310. It also had a sum of theoretical plates, asymmetry (≤1.3) within the acceptable range, and a cumulative desirability of 0.3651. While optimum B provided a slightly better resolution, optimum A was selected for its short analysis time, low energy requirement, and strength in routine laboratory conditions.
At optimum A, the confirmatory runs validated the predicted model outputs. Separation of telmisartan, amlodipine and chlorthalidone with resolution always above 5.0 was reproduced and symmetry factors were within limits. Therefore, the final conditions were those for optimum A, which balanced the analysis performance, robustness, and sustainability for reliable quantification of the combination dosage form.
Results from graphical optimization indicated that flow rate and pH exerted predominant influence, demonstrating the best optimization results at elevated flow rates with adjusted temperatures. The method was thus subsequently finalized employing 35% (v/v) ethanol at an aqueous phase pH of 2.30, with a 1.6 mL min−1 flow rate and 25 °C column oven temperature. Separation of telmisartan, amlodipine and chlorthalidone was achieved reproducibly, with system suitability indicating total resolution greater than 5.0 and asymmetry factors of 1.1–1.3, thus satisfying the criteria of the respective pharmacopeia.
The QbD-driven approach effectively recognized key method variables and characterized a solid design space, enabling dependable and sustainable separation of the combined dosage form. As outlined in the operational matrix, multiple trials were conducted for optimization (Table 2).
| Run no. | Pump flow rate (mL min−1) | Oven temperature (°C) | pH |
|---|---|---|---|
| Conditioning_Run_1 | 1.500 | 25.0 | 2.30 |
| 1 | 1.200 | 25.0 | 2.30 |
| 2 | 1.700 | 25.0 | 2.30 |
| Conditioning_Run_2 | 1.500 | 25.0 | 2.40 |
| 3 | 1.500 | 25.0 | 2.40 |
| Conditioning_Run_3 | 1.500 | 25.0 | 2.50 |
| 4 | 1.200 | 25.0 | 2.50 |
| 5 | 1.700 | 25.0 | 2.50 |
| Conditioning_Run_4 | 1.500 | 30.0 | 2.30 |
| 6 | 1.500 | 30.0 | 2.30 |
| Conditioning_Run_5 | 1.500 | 30.0 | 2.40 |
| 7 | 1.200 | 30.0 | 2.40 |
| 8 | 1.700 | 30.0 | 2.40 |
| 9 | 1.500 | 30.0 | 2.40 |
| 10 | 1.500 | 30.0 | 2.40 |
| 11 | 1.500 | 30.0 | 2.40 |
| Conditioning_Run_6 | 1.500 | 30.0 | 2.50 |
| 12 | 1.500 | 30.0 | 2.50 |
| Conditioning_Run_7 | 1.500 | 40.0 | 2.30 |
| 13 | 1.200 | 40.0 | 2.30 |
| 14 | 1.700 | 40.0 | 2.30 |
| Conditioning_Run_8 | 1.500 | 40.0 | 2.40 |
| 15 | 1.500 | 40.0 | 2.40 |
| Conditioning_Run_9 | 1.500 | 40.0 | 2.50 |
| 16 | 1.200 | 40.0 | 2.50 |
| 17 | 1.700 | 40.0 | 2.50 |
| Conditioning_Run_10 | 1.500 | 40.0 | 2.50 |
Central composite design (CCD) statistical analysis
Three independent variables were optimized using the CCD: the mobile phase pH (C, 2.3–2.5), the column oven temperature (B, 25–40 °C), and the pump flow rate (A, 1.2–1.7 mL min−1). To assess the main, quadratic, and interaction effects, a total of 17 experimental runs—including three center points—were produced. With a G-efficiency of 70.1 (>50% acceptance) and an average predicted variance of 10.2 (<17), the model showed high reliability, confirming the suitability of the design.With R2 = 0.9999 and adj. R2 = 0.9999, the regression analysis of the Rs-map responses showed an excellent model fit, indicating almost perfect agreement between the experimental and predicted values. With a standard error of ±0.017 and a residual mean square error of 0.0003, the experimental data appeared to be highly precise and variable.
Variance inflation factor (VIF) values for all model terms were close to unity (1.00–1.58), confirming the absence of multicollinearity among independent variables. Scaled prediction variance analysis indicated the smallest value at the center point (3.11) and the largest at the axial runs (14.26), which is consistent with the CCD geometry.
Among the studied parameters, flow rate (A) and pH (C) were identified as the most influential factors, showing strong effects on resolution and peak symmetry. Oven temperature (B) also significantly contributed to retention reproducibility. Interaction terms (A × C, B × C, A × B) demonstrated meaningful influence, particularly the combined effect of flow rate and pH, highlighting the necessity of multidimensional optimization. Quadratic terms (A2, B2, C2) were significant, indicating non-linear responses within the studied design space.
A summary is shown in Table 3.
| Parameter | Value | Acceptance criteria | Interpretation |
|---|---|---|---|
| Number of runs (total) | 17 | — | Included 3 center points, 14 non-center points |
| Design type | Response surface – central composite (quadratic) | — | Adequate for multidimensional optimization |
| G-Efficiency | 70.1 | >50% | Indicates good design efficiency |
| Average predicted variance | 10.2 | <17 | Confirms adequate prediction capability |
| R 2 | 0.9999 | >0.90 | Excellent model fit |
| Adjusted R2 | 0.9999 | >0.90 | Strong agreement between predicted and observed values |
| Residual mean square error (MSE) | 0.0003 | As low as possible | Negligible residual error |
| Standard error | ±0.017 | <0.05 desirable | High precision of responses |
| Variance inflation factor (VIF) | 1.00–1.58 | <10 | No multicollinearity observed |
| Scaled prediction variance range | 3.11–14.26 | — | Minimum at center points, maximum at axial runs |
| Significant model terms | A (flow rate), C (pH), A2, B2, C2, A × C, B × C | — | Flow rate and pH most influential |
| p-Value (overall) | <0.0001 | <0.05 | The quadratic model is highly significant |
3D response surface plots show the flow rate and oven temperature impact, and are shown in Fig. 6. The theoretical plates on the left show that efficiency increases at moderate flow and optimized temperature (25–32 °C), but decreases at higher flow and extreme temperature. The asymmetry factor on the right shows that deviation from ideal symmetry (1.0) is minimal around moderate flow (1.2–1.7 mL min−1) and with controlled temperature, while higher flow and higher temperature increase peak tailing.
 | ||
| Fig. 6 3D response: flow rate and temp vs. theoretical plates (left). 3D response: flow rate and temp vs. asymmetry (right). | ||
Overall, the CCD approach successfully established a robust analytical design space, providing a predictive model capable of guiding method conditions with high precision and reliability. The AQbD-based method can be easily transferred to other laboratories by following a standard method-transfer protocol that includes system suitability testing, precision checks and comparison of results between sites. The method was developed using the AQbD approach, and it has a well-defined design space and high robustness. This ensures consistent performance across different instruments and analysts, minimizes transfer failures, and makes the method suitable for routine QC and multi-site industrial use.
The 3D-response surface in Fig. 7A illustrates the interactive influence of flow rate and column temperature on peak asymmetry. As indicated by the curved surface topology, both factors significantly affected the analyte elution profile. At lower flow rates combined with moderate temperatures, the asymmetry values approached unity, indicating optimal peak shape. In contrast, higher flow rates or extreme temperatures produced pronounced peak tailing. The surface curvature confirms a significant interaction between the two factors, consistent with the quadratic model established in the experimental design. In Fig. 7B, showing the 2D-surface response, closely spaced contour lines reflect regions of greater sensitivity to changes in operating conditions, whereas wider spacing indicates stable chromatographic behaviour. In Fig. 7C the design space is constructed by overlaying the response surfaces of critical method attributes, including asymmetry, theoretical plates and retention behaviour. The shaded region represents the multidimensional combination of flow rate and temperature that ensures all responses remain within their predefined acceptance criteria. This proven acceptable range (PAR) defines the operational flexibility of the method under AQbD principles. Operating within this space ensures consistent system suitability and method robustness during routine analysis, and Fig. 7D illustrates the three-dimensional response surface at the optimized mobile phase pH of 2.3, showing the combined effect of flow rate and column temperature on the selected chromatographic response. At the specific flow rate and temperature within the design space, the surface exhibits a smooth plateau, indicating minimal interaction and optimal peak shape. This confirms that the chosen operational conditions provide robust and reproducible chromatographic performance, ensuring that system suitability criteria such as asymmetry and resolution are consistently met under routine analysis.
 | ||
| Fig. 7 (A) 3D surface response. (B) 2D surface response. (C) Design space. (D) Representative 3D surface response of AQbD plots at pH 2.3. | ||
Analytical method validation
Analytical method validation was performed as per Q2 ICH requirements. The observed results are mentioned below.System suitability criteria
System suitability results were determined with different theoretical plates, tailing factor and percent rsd determination for each analyte. The results were within limits (refer to Table 4 for details).| Sr. no. | Chlorthalidone | Telmisartan | Amlodipine |
|---|---|---|---|
| 1 | 101.7 | 97.9 | 100.2 |
| 2 | 102.4 | 98.3 | 101.8 |
| 3 | 102.1 | 99.4 | 101.1 |
| 4 | 102.1 | 99.1 | 101.0 |
| 5 | 103.4 | 98.3 | 102.7 |
| 6 | 102.6 | 99.3 | 100.7 |
| Mean area (n = 6) | 102.4 | 98.7 | 101.3 |
| % RSD | 0.57 | 0.64 | 0.87 |
| Mean asymmetry (n = 6) | 1.11 | 1.16 | 1.10 |
| Resolution | NA | 3.15 | 6.58 |
Specificity
The method effectively differentiates and quantifies the analyte and matrix components. Interference and peak purity were evaluated, with results within limits. The results for the specificity parameters are given in Table 5.| Solution name | Retention time | Peak purity (limit: ≥950) |
|---|---|---|
| Reference solution | ||
| Chlorthalidone | 1.6 | Complies |
| Telmisartan | 4.3 | Complies |
| Amlodipine | 6.2 | Complies |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Test solution | ||
| Chlorthalidone | 1.6 | Complies |
| Telmisartan | 4.3 | Complies |
| Amlodipine | 6.2 | Complies |
Linearity and range
Five linearity levels in triplicate, ranging from 50% to 150% of the working level were evaluated, and then correlation coefficients were calculated as shown in Table 6. Linearity curves are shown in Fig. 8–10.| Parameter | Chlorthalidone | Telmisartan | Amlodipine |
|---|---|---|---|
| Linearity range (µg mL−1) | 25.23–75.69 | 80.11–240.32 | 9.83–29.49 |
| Slope | 14.7155 | 37.2831 | 29.4184 |
| Y-intercept | −14.4871 | −47.8569 | −15.8865 |
| Correlation coefficient | 1.000 | 1.000 | 1.000 |
| Standard deviation of residuals (Sy/x) | 174.6309 | 3654.3617 | 90.0375 |
Accuracy
Accuracy determination was performed in triplicate at three levels of 50%, 100% and 150% with respect to the working level. For all analytes, the results were found to be within limits. The results are shown in Table 7.| Level | Parameter | Chlorthalidone | Telmisartan | Amlodipine |
|---|---|---|---|---|
| 50% | % Mean recovery | 101.2 | 100.0 | 98.1 |
| % RSD | 0.69 | 0.39 | 0.57 | |
| 100% | % Mean recovery | 101.1 | 100.2 | 101.1 |
| % RSD | 0.38 | 0.25 | 0.52 | |
| 150% | % Mean recovery | 101.9 | 100.1 | 101.9 |
| % RSD | 0.64 | 0.65 | 0.66 |
Precision and intermediate precision
The repeatability results were found to be within the limits, ensuring the method's consistency. For detailed results, refer to Table 8.| Sr. no. | Chlorthalidone | Telmisartan | Amlodipine |
|---|---|---|---|
| As such MP (n = 6) | 102.4 | 98.7 | 101.3 |
| As such IP (n = 6) | 101 | 100.4 | 99.5 |
| Absolute difference between MP and IP | 1.4 | 1.7 | 1.8 |
Robustness
Robustness was evaluated with deliberate changes in method parameters, and the results were within limits, confirming procedure reliability. See Table 9 for detailed results.| Conditions | Chlorthalidone | Telmisartan | Amlodipine |
|---|---|---|---|
| Flow 1.4 mL | 100.7 | 100.4 | 100.1 |
| Flow 1.8 mL | 101.0 | 100.6 | 100.2 |
| Col temp 23 °C | 100.7 | 100.3 | 99.7 |
| Col temp 27 °C | 100.6 | 100.3 | 100.1 |
Sample filter study
A sample filter study was performed with a comparison of the centrifuge sample and 0.45 micron filters. The results showed no significant effect on assay values, confirming the filter's suitability for routine analysis. The observed results are tabulated in Table 10.| Sr. no. | Chlorthalidone | Telmisartan | Amlodipine |
|---|---|---|---|
| Centrifuged sample | 102.7 | 98.1 | 100.1 |
| 0.45 µ PVDF filtered | 103.9 | 97.9 | 100.2 |
| 0.45 µ nylon filtered | 103.4 | 97.6 | 100.9 |
Solution stability
Analysis was conducted at different time intervals, evaluating the % assay of the analyte. The results confirmed stability for specific times.Forced-degradation study
Forced degradation performed to check the method's ability to effectively separate degradation peaks from the main compound peak confirms its specificity. Chlorthalidone exhibited the highest degradation under harsh acidic conditions with 80 °C heat, showing 71.5% degradation with a peak purity index of >0.998, confirming the absence of co-eluting impurities. Moderate degradation was observed with base (11.6%) and oxidative stress (8.7%), while thermal and photolytic conditions produced minimal changes (<2%) with peak purity consistently >0.999. Telmisartan remained largely stable under all stress conditions, displaying only 3.4% oxidative degradation and <1% degradation in acid, base, thermal, and photolytic studies, all with peak purity values >0.998, confirming specificity. Amlodipine showed characteristic alkaline and oxidative instability, with 15.3% degradation with base and 9.2% with peroxide, whereas acid, light, and heat resulted in <3% degradation; all degradant peaks and main drug peaks demonstrated purity values >0.999. The clear separation of all degradant peaks from the parent drugs, along with consistently high peak purity indices across all stress conditions, unequivocally confirms that the developed RP-HPLC method is stability-indicating, capable of accurately detecting, resolving, and quantifying each drug in the presence of its degradation products. Percent degradation and peak purity results are shown in Table 11 and Fig. 11.| Conditions | Drug | Treatment applied | Degraded assay (%) | % Degradation | Peak purity |
|---|---|---|---|---|---|
| As such | Chlorthalidone | 96.4 | — | — | |
| Thermal | 72 h heating in a vacuum oven at 110 °C | 96.0 | 0.4 | 999 | |
| Photolytic | 1.2 Million lux h and 200 watt hours meter−1 square light | 96.5 | 0 | 998 | |
| 0.1 N HCl | Heated at 80 °C for 1 h | 24.9 | 71.5 | 999 | |
| 0.1 N NaOH | Heated at 80 °C for 1 h | 84.8 | 11.6 | 999 | |
| 30% H2O2 | Heated at 80 °C for 1 h | 87.7 | 8.7 | 999 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| As such | Telmisartan | 102 | — | — | |
| Thermal | 72 h heating in a vacuum oven at 110 °C | 102.4 | 0 | 999 | |
| Photolytic | 1.2 million lux h and 200 watt hours meter−1 square light | 101.8 | 0.2 | 999 | |
| 0.1 N HCl | Heated at 80 °C for 1 h | 101.2 | 0.8 | 999 | |
| 0.1 N NaOH | Heated at 80 °C for 1 h | 102.75 | 0 | 999 | |
| 30% H2O2 | Heated at 80 °C for 1 h | 98.63 | 3.37 | 1000 | |
| As such | Amlodipine | 97.4 | — | — | |
| Thermal | 72 h heating in a vacuum oven at 110 °C | 95.5 | 1.9 | 991 | |
| Photolytic | 1.2 Million lux h and 200 watt hours meter−1 square light | 103.6 | 0 | 979 | |
| 0.1 N HCl | Heated at 80 °C for 1 h | 101.4 | 0 | 990 | |
| 0.1 N NaOH | Heated at 80 °C for 1 h | 82.1 | 15.3 | 989 | |
| 30% H2O2 | Heated at 80 °C for 1 h | 88.2 | 9.2 | 979 | |
An overall summary of the validation parameters for all active content is given in Table 12.
| Sr. no. | Parameter | Chlorthalidone | Telmisartan | Amlodipine | Acceptance criteria |
|---|---|---|---|---|---|
| 1 | System suitability | % RSD: 0.57; tailing: 1.11 | % RSD: 0.64; tailing: 1.16; resolution: 3.15 | % RSD: 0.87; tailing: 1.10; resolution: 6.58 | % RSD ≤ 2.0% tailing factor ≤ 2.0 resolution ≥ 2.0 |
| 2 | Specificity | Complies (purity ≥ 950) | Complies (purity ≥ 950) | Complies (purity ≥ 950) | No interfering peaks from blank, diluent, placebo peak purity ≥ 950 |
| 3 | Linearity and range | Range: 25.23–75.69 µg mL; r = 1.000 | Range: 80.11–240.32 µg mL; r = 1.000 | Range: 9.83–29.49 µg mL; r = 1.000 | Correlation coefficient (r) ≥ 0.999 linear range: 50–150% of target |
| 4 | Accuracy | Overall mean recovery = 101.4% | Overall mean recovery = 100.1% | Overall mean recovery = 100.4% | Mean recovery 98–102% |
| 5 | Method precision | MP: 102.4%; RSD = 0.57 | MP: 98.7%; RSD = 0.64 | MP: 101.3%; RSD = 0.87 | Repeatability: RSD ≤ 2% (n ≥ 6) |
| 6 | Intermediate precision | IP: 101%; RSD = 0.29 | IP: 100.4%; RSD = 0.18 | IP: 99.5%; RSD = 1.54 | Repeatability: RSD ≤ 2% (n ≥ 6) |
| 7 | Difference between method and intermediate precision | Δ = 1.4% | Δ = 1.7% | Δ = 1.8% | Difference (Δ) < 2% absolute |
| 8 | Robustness | Flow 1.4 mL = 1.7% | Flow 1.4 mL = 1.7% | Flow 1.4 mL = 1.2% | % Change with respect to active contents ≤2% with respect to initial |
| Flow 1.8 mL = 1.4% | Flow 1.8 mL = 1.9% | Flow 1.8 mL = 1.1% | |||
| Col. temp. 23 °C = 1.7% | Col. temp. 23 °C = 1.6% | Col. temp. 23 °C = 1.6% | |||
| Col. temp. 23 °C = 1.8% | Col. temp. 23 °C = 1.6% | Col. temp. 23 °C = 1.2% | |||
| 9 | Filter study | 1.2% | 0.2% | 0.1% | % Difference between filtered vs. centrifuged solution ≤2% |
| 10 | Solution stability | Stable up to 12 h | Stable up to 12 h | Stable up to 12 h | % Change ≤2% from initial results |
Assessment of greenness by green metric tools
The environmental performance of the optimized HPLC method was systematically assessed using multiple green chemistry evaluation tools.A score of 0.75 on the AGREE tool shows that the method aligns with most principles of green analytical chemistry (GAC) with good sustainability. MoGAPI also scored 79, which indicates good greenness, with room to improve. The greenness of the method was also affirmed with a score of 77.5 on the BAGI tool, which shows a good balance of analytical performance and environmental responsibility. The AGREE-Prep tool also showed consistency with green sample preparation, scoring 0.78. The RAPI assessment showed room for improvement, scoring 55.0, which was attributed to energy-intensive operating conditions, longer equilibration times, and moderate solvent use. The ACS Green Chemistry Institute scored the method at 80.96, reflecting excellent alignment to the principles of green chemistry with particular emphasis on solvent safety and low toxicology. Overall, as demonstrated in Fig. 12 and Table 13, the method is environmentally responsible, and developed with excellent to good performance for most metrics of greenness. There are however, moderate issues related to energy and instruments.
| Sr. no. | Green metric | Score | Expanded interpretation |
|---|---|---|---|
| 1 | AGREE | 0.75 | Indicates strong compliance with the 12 principles of green analytical chemistry (GAC). A score of 0.75 reflects that the method uses safer solvents, generates lower waste, avoids hazardous reagents, and employs energy-efficient conditions. Overall, the method demonstrates high sustainability with only minor aspects requiring refinement |
| 2 | MoGAPI | 79 | Reflects good overall greenness across multiple analytical stages, including sample preparation, instrumentation, solvent choice, and waste management. The score of 79 suggests that the method is environmentally responsible, with minor opportunities to further optimize solvent consumption and energy usage |
| 3 | BAGI | 77.5 | Demonstrates a strong balance between analytical performance (precision, robustness, and accuracy) and environmental responsibility. A score of 77.5 shows that the method successfully integrates operational efficiency with greener practices, making it suitable for routine laboratories seeking sustainable workflows |
| 4 | AGREE-Prep | 0.78 | Shows good adherence to green principles specifically related to sample preparation. A score of 0.78 indicates reduced sample-handling steps, minimized reagent use, and lower generation of preparation-related waste, supporting overall method greenness |
| 5 | RAPI | 55 | Indicates moderate greenness. The score is primarily influenced by relatively energy-intensive operating conditions, longer equilibration times, and moderate solvent consumption. While acceptable, this metric highlights potential areas for improvement such as reducing instrument idle time, optimizing temperature settings, or further minimizing solvent usage |
| 6 | ACS Green Chemistry | 80.96 | Reflects excellent alignment with the broader principles of green chemistry, especially due to the use of ethanol (a low-toxicity, renewable, and biodegradable solvent) and minimized hazardous waste. The high score confirms strong environmental performance and reduced human health risks associated with the method |
Conclusion
This study exemplifies the synergistic effect of analytical quality by design (AQbD) and green analytical chemistry in the development of a sustainable chromatographic method for the simultaneous determination of chlorthalidone, amlodipine, and telmisartan in fixed-dose formulations in a drug sample. By applying an analytical quality-by-design (AQbD) approach using a central composite design, the method was systematically optimized for critical variables, namely flow rate, pH, and column temperature, ensuring high method robustness and reduced variability. The use of an ethanol–water mobile phase eliminated toxic organic solvents and achieved well-resolved peaks with a reduced run time, thereby significantly enhancing the method’s environmental sustainability. Comprehensive ICH-compliant validation demonstrated excellent linearity, accuracy, precision, and specificity, with forced-degradation studies confirming full resolution of all degradants and peak purity >0.98, establishing the method's stability-indicating capability. Multiple greenness assessment tools (AGREE, MoGAPI, BAGI, AGREE-Prep, RAPI, and AMGS-tool) consistently reflected strong adherence to green analytical principles, highlighting the method as an environmentally responsible alternative to conventional HPLC approaches. The AQbD-optimized method can also be adapted to real samples, and for single or combined drug samples in the pharmaceutical industry and research organizations working on drug discovery and development.Author contributions
Sudhakar Alave: conceptualization, data curation, formal analysis, investigation, methodology, validation, writing original draft, writing – review and editing. Dr Varsha V. Thorat: writing – review, editing and supervision. Vijay Bagul: writing – review and editing.Conflicts of interest
There are no conflicts of interest to declare.Data availability
The data supporting this article have been included as part of the Supplementary information (SI). Supplementary information: tables, figures and further experimental details. See DOI: https://doi.org/10.1039/d5su00829h.Acknowledgements
The author expresses heartfelt gratitude to Dr Varsha Vishnupant Thorat, the research mentor, for her invaluable guidance, and to Mr Vijay Arjun Bagul for his dedicated technical support throughout the research. Special appreciation is given to ChemClues Lifescience Pvt Ltd for their essential resources that contributed to the study.References
- A. Das, M. Tiwaskar, J. Abdullakutty and A. Pande, J. Assoc. Physicians India, 2024, 72(11), 16–20 CrossRef.
- DrugBank, Amlodipine, https://go.drugbank.com/drugs/DB00381, accessed 18 July 2025 Search PubMed.
- DrugBank, Telmisartan, https://go.drugbank.com/drugs/DB00966, accessed 18 July 2025 Search PubMed.
- DrugBank, Chlorthalidone, https://go.drugbank.com/drugs/DB00310, accessed 18 July 2025 Search PubMed.
- M. W. Nassar, A. Serag and M. A. Hasan, et al. , Sci. Rep., 2025, 15, 25470, DOI:10.1038/s41598-025-09904-0.
- K. P. Kannaiah, A. Sugumaran and H. K. Chanduluru, Microchem. J., 2023, 184(A), 108166, DOI:10.1016/j.microc.2022.108166.
- H. Shamshad and A. Z. Mirza, Futur. J. Pharm. Sci., 2021, 7, 117, DOI:10.1186/s43094-021-00270-y.
- C. S. Funari, R. L. Carneiro, A. J. Cavalheiro and E. F. Hilder, J. Chromatogr. A, 2014, 1354, 34–42, DOI:10.1016/j.chroma.2014.05.018.
- B. Pazhayattil, S. Sharma, J. P. Philip and M. Gischewski-Silva, in Technology Transfer, ed. M. Ingram, Springer, Cham, 2023, vol. 10, ch. 2, DOI:10.1007/978-3-031-32192-4_2.
- M. Eberle, J. T. Wasylenko, D. Kostelac, S. Kiehna, A. Schellinger, Z. Zhang and J. D. Ehrick, Anal. Chem., 2025, 97(1), 12–21, DOI:10.1021/acs.analchem.4c04521.
- International Council for Harmonisation, ICH Q14: Analytical Procedure Development, 2023 Search PubMed.
- K. Sathuluri, R. Bakam and R. Jain, et al. , Accredit. Qual. Assur., 2025, 30, 1–14, DOI:10.1007/s00769-024-01587-w.
- International Council for Harmonisation, ICH Q2(R1): Validation of Analytical Procedures – Text and Methodology, 2005, accessed 1 December 2024, https://www.ich.org/page/quality-guidelines Search PubMed.
- A. Bairagi, R. Kothrukar and H. Chikhale, et al. , Futur. J. Pharm. Sci., 2024, 10, 138, DOI:10.1186/s43094-024-00706-1.
- P. Usgaonkar, A. Adhyapak and R. Koli, Sep. Sci. Plus, 2024, 7(11), e202400155, DOI:10.1002/sscp.202400155.
- S. Armenta, S. Garrigues and M. de la Guardia, TrAC, Trends Anal. Chem., 2008, 27(6), 497–511, DOI:10.1016/j.trac.2008.05.003.
- J. Płotka-Wasylka, Talanta, 2018, 181, 204–209, DOI:10.1016/j.talanta.2018.01.013.
- S. I. Kaya, G. Ozcelikay Akyildiz and S. A. Ozkan, Green Anal. Chem., 2024, 11, 100159, DOI:10.1016/j.greeac.2024.100159.
- M. B. Hicks, W. P. Farrell and C. M. Aurigemma, Green Chem., 2019, 21, 7, 10.1039/C8GC03875A.
- W. Wojnowski, M. Tobiszewski, F. Pena Pereira and E. Psillakis, TrAC, Trends Anal. Chem., 2022, 149, 116553 Search PubMed.
- F. Pena Pereira, W. Wojnowski and M. Tobiszewski, Anal. Chem., 2020, 92, 10076–10082, DOI:10.1021/acs.analchem.0c01887.
- J. Płotka-Wasylka and W. Wojnowski, Green Chem., 2021, 23, 1–10, 10.1039/D1GC02318G.
- P. M. Nowak, W. Wojnowski, N. Manousi, V. Samanidou and J. Płotka-Wasylka, Green Chem., 2025, 27, 5546, 10.1039/D4GC05298F.
- R. P. Mistry, C. Shah and R. Jat, Folia Med., 2022, 64, 103–109, DOI:10.3897/folmed.64.e64339.
- R. L. C. Sasidhar, S. Vidyadhara, B. Deepti, K. Tejaswi and J. Suhasini, Orient. J. Chem., 2014, 30(4) DOI:10.13005/ojc/300442.
- K. Kalyani, V. Anuradha, S. Vidyadhara, R. L. C. Sasidhar and T. N. V. G. Kumar, Orient. J. Chem., 2016, 32(3), 1401–1408, DOI:10.13005/ojc/320340.
- M. Kumar, et al. , Int. J. Pharm. Sci. Drug Res., 2021, 13, 288–294, DOI:10.25004/IJPSDR.2021.130308.
- H. T. T. Nguyen, T. V. Duong and D. T. Phan, Med. Pharm. Res., 2023, 7, 67–74, DOI:10.32895/UMP.MPR.7.4.8.
- P. Prajapati, A. Patel and S. Shah, J. Chromatogr. Sci., 2023, 61, 160–171, DOI:10.1093/chromsci/bmac030.
- M. A. Kamel, C. K. Nessim, A. M. Michael, S. S. Abbas and H. M. Marzouk, BMC Chem., 2024, 18, 166, DOI:10.1186/s13065-024-01276-2.
- P. Kulkarni and D. Gangrade, Int. J. Pharm. Sci. Res., 2017, 8(1), 268–276, DOI:10.13040/IJPSR.0975-8232.8(1).268-76.
- G. H. Patel, S. D. Adeshra and D. B. Meshram, Int. J. Pharm. Drug Anal., 2021, 9(3), 39–47, DOI:10.47957/ijpda.v9i3.480.
- S. Pawar, P. K. Kusuma, B. Bhadru, S. T. Shanmugam, R. K. Kumarachari, B. R. Narapureddy and H. Onohuean, Acta Chromatogr., 2025 DOI:10.1556/1326.2025.01374.
- S. Hingde and S. Boddu, World J. Pharm. Res., 2017, 6(6), 936–945, DOI:10.20959/wjpr20176-8533.
- M. Panda, V. Dadi, S. R. Yarraguntla and V. P. R. K. Rao, Res. J. Pharm. Technol., 2023, 16(2), 509–513, DOI:10.52711/0974-360X.2023.00086.
- S. A. Kanchana, A. Aruna, V. Niraimathi and A. J. Suresh, Res. J. Pharm. Technol., 2011, 4, 428–429 Search PubMed.
- A. D. Dhumal and G. Barabde, Int. J. Pharm. Sci. Res., 2024, 15(8), 2541–2550, DOI:10.13040/IJPSR.0975-8232.15(8).2541-50.
- S. Nalwade, V. R. Reddy, D. D. Rao and I. K. Rao, Sci. Pharm., 2011, 79, 69–84, DOI:10.3797/scipharm.1006-10.
- M. Attimarad, K. N. Venugopala, N. Sreeharsha, M. S. Chohan, S. Shafi, A. B. Nair and S. Pottathil, Separations, 2021, 8, 86, DOI:10.3390/separations8060086.
- S. N. Atapattu, J. Chromatogr. Open, 2024, 5, 100135, DOI:10.1016/j.jcoa.2024.100135.
- T. Yi, L. Zhu, W.-L. Peng, X.-C. He, H.-L. Chen, J. Li, T. Yu, Z.-T. Liang, Z.-Z. Zhao and H.-B. Chen, LWT–Food Sci. Technol., 2015, 62, 194–201, DOI:10.1016/j.lwt.2015.01.003.
- T. Yi, H. Zhang and Z. Cai, Phytochem. Anal., 2007, 18, 387–392, DOI:10.1002/pca.993.
- J. Feng, H. Ren, Q. Gou, L. Zhu, H. Ji and T. Yi, Anal. Methods, 2016, 8, 1557–1564, 10.1039/C5AY02941D.
- T. Yi, K. S.-Y. Leung, G.-H. Lu, K. Chan and H. Zhang, Chem. Pharm. Bull., 2006, 54, 255–259, DOI:10.1248/cpb.54.255.
- T. Yi, K. S.-Y. Leung, G.-H. Lu, H. Zhang and K. Chan, Chem. Pharm. Bull., 2005, 53, 1480–1483, DOI:10.1248/cpb.53.1480.
- International Council for Harmonisation, ICH Q1A(R2): Stability Testing of New Drug Substances and Products, 2005, accessed 1 December 2024, https://www.ich.org/page/quality Search PubMed.
Footnote |
| † The method was developed using an analytical quality by design (AQbD) and green analytical approach, ensuring robustness, precision, and minimal environmental impact through the use of ethanol–water as an eco-friendly mobile phase. |
| This journal is © The Royal Society of Chemistry 2026 |