Open Access Article
Open Access ArticleA novel method to screen biodegradability for the early assessment of cellulosic rheology modifiers
Moumita
Bhaumik†
,
Chiranjeevi
Thulluri†
,
Arindam
Roy
and
Harshad
Ravindra Velankar
 *
*
Alt Material Innovations Private Limited (short form ‘altM’), Plot No. 35, Electronic City, Phase 2, Industrial Area, Konappana Agrahara, Bengaluru, Karnataka 560100, India. E-mail: harshad@altm.bio
First published on 27th November 2025
Abstract
In this study, we developed a new method for early-stage biodegradability assessment of cellulosic rheology modifiers (CRMs). Viscosity reduction was used as the primary indicator of polymer degradation. Complementary analyses included molecular weight changes (gel permeation chromatography, GPC), total carbohydrate content (TCC), and chemical oxygen demand (COD). Mixed microbial consortia from environmental sources ensured ecologically relevant conditions. Five CRMs including HPC-J (hydroxypropyl cellulose, J type), HPC-M (hydroxypropyl cellulose, M type), HPMC (hydroxypropyl methyl cellulose), HEMC (hydroxyethyl methyl cellulose), and cet-HEC (cetyl hydroxyethyl cellulose) were monitored over 8 weeks. Molecular weight dropped significantly, particularly for HPMC, which exhibited a 46.1-fold decrease, confirming chain scission. TCC declined sharply, with HPC-J surpassing an 85% reduction by day 56, evidencing microbial uptake. Furthermore, a predictive mathematical model was established, revealing the degradation sensitivity factor (‘a’), which ranged from a = 0.48 (for the highly resistant HPMC) to a = 4.85 (for the extremely sensitive cet-HEC). This simple, low-cost approach enables simultaneous small-scale testing as an early biodegradability screen, offering a practical decision tool before moving to standardized protocols and helping identify structural modifications that may hinder microbial breakdown.
Sustainability spotlightThis study presents a simple, low-cost biodegradability screening method for cellulose-based rheology modifiers (CRMs) that uses viscosity loss, molecular weight decrease, and total carbohydrate reduction as early indicators of microbial degradation. By enabling rapid identification of structural features that slow or prevent biodegradation, the approach supports the design of truly biodegradable CRMs at the R&D stage, well before resource-intensive OECD/ISO testing. Because CRMs are widely used in cosmetics and personal care products that ultimately enter wastewater and soils, early screening can help reduce persistent polymer residues, lower microplastic formation risk, and accelerate the transition to safer, more sustainable consumer materials. |
1 Introduction
The growing use of polymers in cosmetics and consumer products has intensified concerns regarding their persistence in the environment. To prevent long-term ecological impact, such molecules must undergo complete biodegradation rather than partial fragmentation into microplastics.1 Achieving full mineralization is essential to mitigate the accumulation of polymer residues in wastewater and soil. Consequently, there is increasing momentum toward replacing conventional petroleum-derived polymers with biopolymers produced from renewable resources.2 Within this sustainability transition, rheology modifiers (RMs) represent a particularly important yet under-examined class of functional polymers.Typically, RMs are incorporated into liquid systems to control key properties such as viscosity, stability, suspension, and spreadability.3,4 They are widely used across diverse industries, including personal care, paints and coatings, construction materials, and pharmaceuticals.5–7 Depending on their source, RMs can be classified as natural (e.g., xanthan gum), synthetic (e.g., polyacrylates), or semi-synthetic.8 Our study focuses on semi-synthetic cellulose-based rheology modifiers (CRMs), which are derived from abundant, renewable cellulose but chemically modified via etherification to enhance solubility, stability, and salt tolerance. Although CRMs retain a natural polymeric backbone, their chemical substitution patterns such as the degree of substitution, type of substituent (hydroxypropyl, methyl, ethyl, and cetyl), and side-chain length strongly influence both their performance and biodegradability.9 Persistent or non-degradable CRMs thus pose emerging sustainability challenges despite their renewable origin.10
Therefore, sustainable polymer design requires the evaluation of biodegradability at the earliest stages of research and development. However, limited guidance exists for conducting such assessments during polymer development. Established industry standards from the OECD (Organisation for Economic Co-operation and Development)10 and ISO (International Organization for Standardization), such as the OECD 301 B/F tests (CO2 evolution tests) and ISO 14851 (ultimate biodegradability of plastics),11 provide robust protocols for determining CO2 evolution and ultimate biodegradability, respectively. However, these methods are time-consuming, resource-intensive, and unsuitable for rapid R&D screening.12,13 They typically require long incubation periods (28 days to several months), specialized equipment such as respirometers, and relatively large sample quantities that are often unavailable during early-stage synthesis. Moreover, they measure only endpoint parameters such as CO2 evolution or O2 consumption that confirm ultimate mineralization10,11 but offer limited mechanistic insight into intermediate degradation steps such as chain scission or microbial assimilation. The absence of such mechanistic understanding constrains the development of structure–biodegradability correlations, which are essential for the rational design of sustainable polymers.14
Considering the above, in this study we developed a low-cost, multi-parametric screening approach that enables rapid and mechanistic evaluation of CRM biodegradability during early development. The method employs viscosity reduction as a functional and sensitive indicator of microbial degradation, complemented by gel permeation chromatography (GPC), total carbohydrate content (TCC), and chemical oxygen demand (COD) analyses to provide a comprehensive degradation profile. Together, these parameters capture the sequential progression of biodegradation from polymer chain scission to microbial uptake and ultimate mineralization to some extent.
More precisely, viscosity reduction serves as the primary signal of degradation, reflecting the loss of chain entanglement and molecular weight as microbial enzymes cleave the polymer backbone. GPC analysis quantitatively verifies molecular weight reduction, confirming the mechanistic basis of functional loss. Concurrently, the depletion of dissolved carbohydrate content (TCC) indicates microbial assimilation of degradation intermediates, while the decline in COD reflects overall mineralization of the organic matter into CO2, water, and biomass. Collectively, these four measurements, functional loss, molecular degradation, microbial uptake, and mineralization, along with predictive mathematical modelling provide an integrated and mechanistic understanding of polymer biodegradation kinetics in real time.
The developed method allows small-scale, ecologically relevant testing using mixed microbial consortia and serves as an effective preliminary R&D tool prior to OECD or ISO testing. In this study, five commercially representative CRMs, hydroxypropyl cellulose (HPC-J and HPC-M), hydroxypropyl methylcellulose (HPMC), hydroxyethyl methylcellulose (HEMC), and cetyl hydroxyethylcellulose (cet-HEC) were evaluated to demonstrate the utility of this approach and to elucidate structural factors governing biodegradability.
2 Materials and methods
2.1 Chemicals and reagents
All chemicals were of analytical grade unless otherwise specified.2.2 Biodegradability screening assays
Biodegradation screening assays were conducted in 50 mL glass tubes, each containing 20 mL of minimal salts medium. All tubes were incubated in a temperature-controlled shaker incubator. The biodegradation experiment comprised three main phases: (a) sample collection and microbial consortium preparation: we chose a mixed microbial consortium for the study because such diverse communities containing bacteria, fungi, and other microbes are generally more effective than single strains at degrading biopolymers in natural environments.15 Their higher effectiveness arises from synergistic interactions since different microbes contribute complementary metabolic pathways, one organism's by-products serve as nutrients for another, and together they provide a wider range of enzymes capable of breaking down complex biopolymer structures.15 Hence, to prepare the mixed microbial consortium, environmental samples were collected from a local sewage pit (12°51′47.1″N 77°40′08.3″E) and a solid waste composting yard (12°51′47.2″N 77°41′10.9″E). Soil samples were taken from the upper soil layer (5–10 cm depth). Approximately 100 g of each soil sample was mixed with 100 mL of deionized water for 30 minutes under ambient conditions and then allowed to settle. The resulting suspension was filtered through a nylon strainer and subsequently centrifuged at 5000 rpm for 5 minutes. The soil extract (10 mL) was then added to 100 mL of sterile nutrient broth in a 250 mL conical flask and incubated at 37 °C for 48 hours to enrich the mixed microbial consortium (achieving an optical density, OD600, of 1.0). Concurrently, liquid samples from the sewage pit were directly used to inoculate a separate batch of nutrient broth. This sewage-inoculated broth was then combined with the soil extract-enriched culture. The resulting mixed microbial culture served as the inoculum for all subsequent biodegradation experiments. (b) Preparation of minimal medium (MM) and test substrates: the standard minimal medium (whose stock solution-A contains K2HPO4, 21.75 g L−1; KH2PO4, 8.5 g L−1; NH4Cl, 0.5 g L−1; Na2HPO4·2H2O, 33.4 g L−1 and stock solutions B, C and D contain 22.5 g L−1 of MgSO4·7H2O, 36.4 g L−1 of CaCl2·2H2O, and 0.25 g L−1 of FeCl3·6H2O, respectively), as described in ISO 14851,11 was used as the basic nutrient medium for the biodegradation assessment of all cellulose-based rheology modifiers (CRMs). The final nutrient medium was prepared by mixing stock solutions listed in Table S5, 11 mL of solution A, 1 mL of solution B, 1 mL of solution C, and 1 mL of solution D, followed by adjusting the final volume to 1000 mL with de-mineralized water. This prepared minimal medium was then distributed into 50 mL test tubes, with 20 mL per tube. Different CRMs viz., HPC-J, HPC-M, HPMC, HEMC, and cet-HEC were added to individual tubes at specific concentrations of 4.4% w/v, 1.8% w/v, 1.5% w/v, 0.8% w/v, and 0.4% w/v, respectively, and dissolved at room temperature. The initial viscosities of the CRMs were in the range of 180–230 cP. After assessing these viscosities, all tubes were autoclaved at 121 °C for 15 minutes and upon cooling to room temperature, samples were vortexed for 30 minutes to ensure uniform dissolution of the CRM constituents. Viscosity was re-analyzed post-sterilization; no reduction occurred due to autoclaving. (c) Setting up of biodegradation tests: for each CRM test substrate, the experimental setup included the following four groups of test tubes, prepared in duplicate sets: positive control (tube 1): 20 mL MM media with 1% w/v glucose + 1 mL inoculum (0.5% v/v). Negative control (tube 2): 20 mL MM media + 1 mL inoculum (0.5% v/v). Abiotic control (tube 3): 20 mL MM media + respective test CRM. Biotic test (tube 4): 20 mL MM media + respective test CRM + 1 mL inoculum (0.5% v/v). Prior to inoculation, the mixed microbial culture (in nutrient broth) was pre-incubated at 37 °C and 150 rpm for 24 hours. A total of five duplicate sets were prepared for each CRM. After inoculation, all experimental test tubes were incubated at 37 °C in a shaker incubator at 150 rpm. Samples were withdrawn from duplicate sets every two weeks for a total duration of eight weeks (56 days), corresponding to sampling points at week 0 (day 0), week 2 (day 14), week 4 (day 28), week 6 (day 42), and week 8 (day 56). These samples were subsequently analyzed for various parameters, including viscosity, polymer molecular weights, total carbohydrate content (TCC), and chemical oxygen demand (COD), to assess and compare the degree of biodegradation of the CRMs.2.3 Analytical methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 464
464![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; 1
000 Da; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130
130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; 249
000 Da; 249![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; 125
000 Da; 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; 34
000 Da; 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 Da; 19
600 Da; 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 Da; 3290 Da; 1220 Da) (SI, S2 and S3).
100 Da; 3290 Da; 1220 Da) (SI, S2 and S3).
| η ∝ Mwa | (1) |
In general, to create a dimensionless, predictive metric independent of the material properties, we define the retention factor (R) for both viscosity and molecular weight.
| Viscosity retention factor (ηR) = ηt/η0 | (2) |
Similarly,
| Molecular weight retention factor (MwR) = Mwt/Mw0 | (3) |
By applying the power law equation (eqn (1)) to the retention factors, we derive the fundamental quantitative model for polymer degradation extent.
| ηR = (Mw,R)a | (4) |
2.3.5.1 Establishing the predictive exponent (a). The critical factor is the exponent ‘a’. By plotting the natural logarithm of the viscosity retention (ln(ηR)) against the natural logarithm of the molecular weight retention (ln(Mw,R)), the slope of the resulting linear fit directly yields the value of ‘a’:
| ln(nR) = a × ln(Mw,R) | (5) |
3 Results and discussion
The use of environmentally derived microbial consortia for assessing the biodegradation of rheology modifiers (RMs) offers a more ecologically relevant and robust alternative to single-strain studies.18 Microbial communities present in sewage, soil, and compost are naturally adapted to break down complex organic substrates, equipped with diverse enzymatic systems and synergistic metabolic interactions that enhance polymer degradation.19,20 These mixed microbial cultures more accurately mimic natural biodegradation processes, where the consortia function collectively through co-metabolism, enzymatic complementation, and quorum sensing to efficiently decompose the organic substances. This approach enhances the predictive reliability of laboratory-based biodegradation assays, ensuring that degradation kinetics and metabolic pathways more closely reflect those occurring under natural environmental conditions. Therefore, as mentioned above, a mixed consortium was developed using the culture samples collected from various environmental sources and used in the current study.3.1 Biodegradation assessment of CRMs via viscosity changes
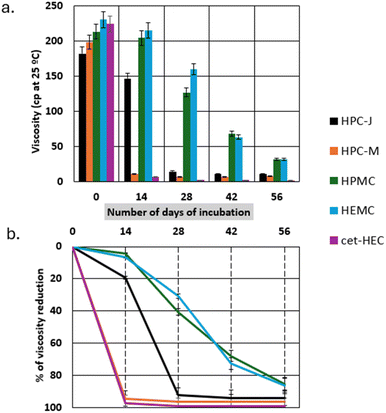
As mentioned in the test protocol, a total of five different types of CRMs (HPC-J & M, HPMC, HEMC, and cet-HEC) were used to assess their biodegradability using the newly developed laboratory protocol. Among the five RMs tested, HPC-J and HPC-M were synthesized in-house, while the others were procured from the market. The results, depicted in Fig. 1, demonstrate significant changes in viscosity for all tested polymers, indicating their susceptibility to biodegradation.Fig. 1a presents the absolute viscosity values (in centipoise, cP) at 25 °C as a function of incubation time while Fig. 1b highlights the percentage viscosity reduction. At day 0, all samples exhibited characteristic viscosities, with HPC-J and HPC-M showing initial viscosities around 180 and 200 cP, HPMC ∼213 cP, HEMC ∼230 cP, and cet-HEC ∼220 cP. As the test progressed, a noticeable reduction in viscosity was observed across all samples. For HPC-M and cet-HEC, the viscosity dropped sharply within the first 14–28 days, reaching near-zero values by day 42. HPC-J also showed a rapid decrease, with its viscosity significantly diminishing by day 14 and approaching near-zero by day 28. Extensive degradation was observed for HPC-J, cet-HEC, and HPC-M, all achieving close to 100% viscosity reduction within 28–42 days (Fig. 1b). Specifically, both HPC-M and cet-HEC showed an exceptionally rapid decline, reaching nearly 90% reduction by day 14. On the other hand, HEMC and HPMC displayed a more linear reduction, achieving ∼80% reduction by the end of the incubation period (8th week). These results consistently indicate a substantial loss of rheological properties, directly correlating with cellulosic polymer degradation.
The developed viscosity-based method offers a distinct advantage over conventional biodegradation assessment techniques such as those based on carbon balance (e.g., OECD 301 B)10 and qualitative material property loss (e.g., tensile strength for plastics, or environmental fate assessments).8 Moreover, traditional methods often require complex instrumentation, extended incubation periods, and intricate analytical procedures (e.g., CO2 evolution monitoring, extensive material characterization, or tracking residual fragments). In contrast, the viscosity measurement is relatively simple, cost-effective, and rapid, providing a direct and instinctive indication of polymer degradation through the loss of its defining rheological property.21 The ‘less effort’ aspect of this method makes it particularly attractive for preliminary screening of biodegradable polymers or for quality control in manufacturing where rapid assessment is valuable.
For rapid assessment of biodegradability of RMs, the correlation between viscosity loss and polymer chain breakdown is very strong. While extensive research worldwide is focused on developing biodegradable biopolymers, progress is often slowed by the lack of a quick screening method. Existing protocols can be costly and time-consuming, making this simple approach especially promising for accelerating the development and evaluation of novel biodegradable rheology modifiers, thereby supporting more sustainable material solutions. Moreover, when interpreted in the context of standardized biodegradability frameworks, the results of biodegradability assessment align partially with OECD criteria for ready or inherent biodegradability particularly for cet-HEC and the in-house HPCs, which showed >60% degradation within 28–42 days. However, the commercial samples, with slower viscosity reduction and incomplete breakdown within 60 days, may not fully meet OECD 301 ‘ready biodegradability’ thresholds, although they may qualify as inherently biodegradable under OECD 302. As ISO and ASTM guidelines do not explicitly define ‘readiness’ but instead use threshold-based pass criteria (e.g., ≥90% CO2 evolution in compost within 180 days), the observed trends suggest that most samples, particularly cet-HEC and HPCs, would likely meet ISO 17088 (ref. 22) or ASTM D5338 (ref. 23) biodegradability criteria under composting conditions. The extended degradation time for commercial products may necessitate longer test durations or modified conditions to fully assess their environmental fate.
3.2 Changes in molecular weight of CRMs during biodegradation
As mentioned above, the biodegradation of all five tested RMs was also analyzed by monitoring changes in their molecular weight properties using Gel Permeation Chromatography (GPC) over the biodegradation period. The weight-average molecular weight (Mw), number-average molecular weight (Mn), peak molecular weight (Mp), z-average molecular weight (Mz), and polydispersity index (PDI) were determined for each sample at different time points. The GPC results clearly demonstrate that the developed biodegradation technique effectively degrades the CRMs, leading to a substantial reduction in their molecular weights.
Table 1 shows that all tested polymers exhibited a notable reduction in their weight-average molecular weight (Mw) over the biodegradation testing incubation period, indicating successful degradation. The observed decrease in Mw across all tested polymers is a direct and strong indicator of polymer chain hydrolysis, which is characteristic of biodegradation processes.24,25 This aligns with the expectation that enzymatic or microbial activity targets the RM's polymeric backbone, breaking down larger macromolecules into smaller fragments.21,26 As can be seen in Table 1, specifically, HPC-J showed a decrease in Mw from an initial 405![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 226 Da (day 0) to 135
226 Da (day 0) to 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 912 Da by the end of the test, representing a 3.0-fold reduction. Similarly, HPC-M demonstrated a significant 4.6-fold reduction in Mw, dropping from 1
912 Da by the end of the test, representing a 3.0-fold reduction. Similarly, HPC-M demonstrated a significant 4.6-fold reduction in Mw, dropping from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 036
036![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 251 Da to 223
251 Da to 223![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596 Da over the same period. Nevertheless, HPMC exhibited the most substantial degradation, with its Mw decreasing from 1
596 Da over the same period. Nevertheless, HPMC exhibited the most substantial degradation, with its Mw decreasing from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 429
429![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529 Da to 30
529 Da to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 996 Da, corresponding to an impressive 46.1-fold reduction after 56 days. HEMC also showed considerable degradation, with Mw decreasing from 1
996 Da, corresponding to an impressive 46.1-fold reduction after 56 days. HEMC also showed considerable degradation, with Mw decreasing from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 343
343![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 Da to 191
380 Da to 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 Da, a 7.0-fold reduction. Lastly, Mw of cet-HEC decreased from 243
737 Da, a 7.0-fold reduction. Lastly, Mw of cet-HEC decreased from 243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 Da to 88
760 Da to 88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936 Da, achieving a 2.7-fold reduction.
936 Da, achieving a 2.7-fold reduction.
| CRM type | Day | Sample | M w (Da) (fold reduction) | M p (Da) | M n (Da) | M z (Da) | PDI (Mw/Mn) |
|---|---|---|---|---|---|---|---|
| a RM: rheology modifier; HPC: hydroxypropyl cellulose; HPMC: hydroxypropyl methyl cellulose; HEMC hydroxyethyl methyl cellulose; cet-HEC: cetyl-hydroxyethyl cellulose; PDI: poly dispersity index. | |||||||
| HPC-J | 0 | Control | 405![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 226 (0.0) 226 (0.0) |
271![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094 094 |
155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 794 794 |
998![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 279 279 |
2.6 |
| 14 | T1 | 146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 515 (2.8) 515 (2.8) |
122![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790 790 |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 876 876 |
244![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 691 691 |
2.6 | |
| 28 | T2 | 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993 (2.9) 993 (2.9) |
118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 885 885 |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182 182 |
140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993 993 |
1.7 | |
| 42 | T3 | 138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 414 (2.9) 414 (2.9) |
121![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390 390 |
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923 923 |
220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 971 971 |
1.7 | |
| 56 | T4 | 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 912 (3.0) 912 (3.0) |
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 291 291 |
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 613 613 |
212![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742 742 |
1.7 | |
| HPC-M | 0 | Control | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 036 036![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 251 (0.0) 251 (0.0) |
797![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 921 921 |
670![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 323 323 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648 648![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 706 706 |
1.5 |
| 14 | T1 | 251![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867 (4.1) 867 (4.1) |
192![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 042 042 |
121![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 634 634 |
520![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 080 |
2.1 | |
| 28 | T2 | 243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 989 (4.2) 989 (4.2) |
178![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 541 541 |
132![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780 780 |
453![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 252 252 |
1.8 | |
| 42 | T3 | 243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 574 (4.3) 574 (4.3) |
187![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339 339 |
124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 613 613 |
471![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 837 837 |
2 | |
| 56 | T4 | 223![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596 (4.6) 596 (4.6) |
175![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 532 532 |
113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840 840 |
428![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 427 427 |
2 | |
| HPMC | 0 | Control | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 429 429![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529 (0.0) 529 (0.0) |
845![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 830 830 |
952![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 147 147 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264 264![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 861 |
1.5 |
| 14 | T1 | 755![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 418 (1.9) 418 (1.9) |
564![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 089 089 |
610![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 210 210 |
953![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 328 328 |
1.2 | |
| 28 | T2 | 159![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 509 (9.0) 509 (9.0) |
146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 049 049 |
93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 054 054 |
252![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 489 489 |
1.7 | |
| 42 | T3 | 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968 (21.0) 968 (21.0) |
58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 985 985 |
48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 595 595 |
91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 166 166 |
1.4 | |
| 56 | T4 | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 996 (46.1) 996 (46.1) |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 593 593 |
26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 716 716 |
36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 177 177 |
1.2 | |
| HEMC | 0 | Control | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 343 343![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 (0.0) 380 (0.0) |
908![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 970 |
288![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958 958 |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 419 419![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 581 581 |
4.7 |
| 14 | T1 | 589![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 677 (2.3) 677 (2.3) |
562![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 665 |
199![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844 844 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 337 337![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860 860 |
3.0 | |
| 28 | T2 | 432![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249 (3.1) 249 (3.1) |
367![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 561 561 |
182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 691 691 |
912![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 427 427 |
2.4 | |
| 42 | T3 | 193![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 459 (6.9) 459 (6.9) |
178![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 184 184 |
107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 107 |
325![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 369 369 |
1.8 | |
| 56 | T4 | 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 737 (7.0) 737 (7.0) |
187![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339 339 |
141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 962 962 |
248![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941 941 |
1.4 | |
| cet-HEC | 0 | Control | 243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 (0.0) 760 (0.0) |
293![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555 555 |
138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895 895 |
355![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 378 378 |
1.75 |
| 14 | T1 | 111![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 604 (2.2) 604 (2.2) |
105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 651 651 |
78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 704 |
147![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
1.42 | |
| 28 | T2 | 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 532 (2.6) 532 (2.6) |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 323 323 |
67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923 923 |
1.39 | |
| 42 | T3 | 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 (2.5) 360 (2.5) |
84![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 369 369 |
68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 970 |
131![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 321 |
1.41 | |
| 56 | T4 | 88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936 (2.7) 936 (2.7) |
76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 744 744 |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 497 497 |
119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 092 |
1.4 | |
The varying degrees of molecular weight reduction among the different RMs highlight differences in their susceptibility to the biodegradation process. During the biodegradation period, HPMC showed the highest steady reduction in Mw (46.1-fold), suggesting that its specific chemical structure or degree of substitution may render it particularly vulnerable to the biodegrading agents in this system. This high susceptibility could be due to more accessible glycosidic linkages or less steric hindrance compared to the other modified cellulose derivatives.27,28 Conversely, HPC-J, HPC-M, and cet-HEC exhibited more reasonable but still significant reductions, indicating their biodegradability, albeit at a slower rate or to a lesser extent under the experimental conditions. Overall, the fold change values of Mw presented in Table 1 serve as a compelling quantitative measure of this degradation efficiency.
In addition, the polydispersity index (PDI), which reflects the breadth of molecular weight distribution, also showed changes for most samples over time, which further supports the interpretation of their biodegradability. For instance, the PDI of HEMC significantly decreased from 4.7 to 1.4 (Table 1), suggesting a narrower distribution of molecular weights as degradation progressed. Similarly, the PDI of HPMC narrowed from 1.5 to 1.2 while HPC-J, HPC-M, and cet-HEC exhibited slight decreases or stability in their PDI values, largely indicating a more uniform degradation process. As discussed earlier, a decrease in PDI of cellulosic polymers during the biodegradation test often signifies that the degradation process preferentially targets the larger polymer chains or that the degradation products converge towards a more uniform size distribution. This could occur if the microbial enzymes involved in the degradation process have specific cleavage sites or if the smaller, more resistant fragments accumulate.28,29 As explained by this mechanism, the initial higher PDI of HEMC (4.7) followed by a significant drop to 1.4 (Table 1) suggests that the biodegradation effectively broke down a very broad initial molecular weight distribution into a much narrower one. For HPC-J, HPC-M, and cet-HEC, the relatively stable or slightly decreasing PDI values, even with significant Mw reduction, suggest a more uniform degradation across the molecular weight distribution or that the remaining polymer fragments maintain a somewhat consistent size range.
3.3 Total carbohydrate analysis during biodegradation assessment of CRMs
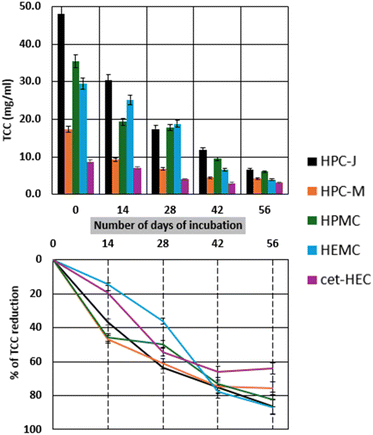
The biodegradation of CRMs was further monitored by determining the residual total carbohydrate concentration (TCC) in the test medium. Fig. 2 illustrates the changes in TCC and the corresponding percentage reduction for each CRM. Fig. 2a displays the absolute TCC (mg mL−1) while Fig. 2b presents the percentage of TCC reduction at different incubation time points, providing a clearer view of the extent of carbohydrate degradation.On day 0, HPC-J had the initial TCC concentration at approximately 48 mg mL−1, followed by HPMC (35.5 mg mL−1) and HEMC (29.5 mg mL−1). HPC-M had a lower initial concentration (17.3 mg mL−1), and cet-HEC had the lowest (8.7 mg mL−1). All samples showed a consistent decrease in TCC concentration as the incubation progressed. Explicitly, HPC-J demonstrated a significant absolute reduction from 48 mg mL−1 at day 0 to <10 mg mL−1 by day 56. Similarly, TCC content of HPMC and HEMC also decreased markedly, falling from their initial 35.5 mg mL−1 and 29.5 mg mL−1 concentrations, respectively, to below 10 mg mL−1. Meanwhile, HPC-M and cet-HEC, starting with lower initial concentrations, also exhibited significant reductions over time, reaching concentrations below 5 mg mL−1 by day 56.
Overall, HPC-J resulted in the highest percentage reduction, approaching ∼90% by day 56. Similarly, HEMC also showed an 86% reduction by the end of the study, while HPMC concentrations reduced by 82% and HPC-M reduced by ∼76%. On the other hand, cet-HEC exhibited a moderate reduction, of ∼65% reduction by day 56. The rapid initial reduction in TCC content for HPC-J, HPMC, HEMC, and HPC-M within the first 14–28 days was consistent with the rapid changes observed in their corresponding molecular weight and viscosities.
Accordingly, the reduction in overall TCC across all CRMs provides strong evidence of their degradation and assimilation during the biodegradation test. Being cellulose derivatives, these polymers are composed primarily of carbohydrate units,30 and their disappearance from the solution indicates that the microbial consortia or enzymes involved in their biodegradation are actively breaking down and consuming these polymeric sugars.31 This quantitative loss of carbohydrate mass correlates with the qualitative and rheological changes observed in the molecular weight and viscosity analyses (Table 1 and Fig. 1).
Moreover, the strong reduction in TC concentration, particularly for HPC-J, HEMC, and HPMC, further reinforces the high biodegradability of these specific polymers. The TCC reduction indirectly correlates to the ‘carbon balance’ aspect considered during conventional biodegradation tests (e.g., OECD, ISO, and ASTM)10,11,22,23 which quantify degradation by measuring the carbon dioxide evolution or loss of substrate. Overall, our method is simpler to execute than full carbon balance tests, and the direct measurement of TCC loss provides considerable evidence of material consumption, serving as a key indicator of CRM's biodegradability.
Considering our results, the varying rates and extent of TCC reduction among the different polymers can be attributed to their unique structural modifications, degrees of substitution or molar substitution, and accessibility of glycosidic bonds, which influence their susceptibility to enzymatic hydrolysis and microbial utilization.30 This total carbohydrate analysis provides a crucial quantitative metric, complementing the rheological and molecular weight data by directly measuring the biodegradation of the CRM substrate. Furthermore, it offers a simpler approach for material mineralization or assimilation, reinforcing the validity of the newly developed biodegradation assessment approach in this study.
3.4 Chemical oxygen demand (COD) analysis during biodegradation assessment
In addition to TCC analysis, the initial and final day chemical oxygen demand (COD) was also measured in the biodegradation test samples to quantify the reduction in total oxidizable organic matter due to CRM degradation (Table 2). A reduction in COD values was observed for all tested CRMs. HPC-J showed a decrease in COD from 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 136 ppm on day 0 to 59
136 ppm on day 0 to 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 ppm on day 56, representing a 16.67% reduction. Correspondingly, HPC-M exhibited a 40% reduction, with COD dropping from 29
280 ppm on day 56, representing a 16.67% reduction. Correspondingly, HPC-M exhibited a 40% reduction, with COD dropping from 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 ppm to 17
640 ppm to 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 784 ppm. Likewise, HPMC also showed a reduction of 40.01%, decreasing from 11
784 ppm. Likewise, HPMC also showed a reduction of 40.01%, decreasing from 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 856 ppm to 7113 ppm. HEMC, while showing a reduction, had the lowest percentage decrease at 9.11%, with COD changing from 6522 ppm to 5928 ppm. Finally, cet-HEC exhibited a 27.78% reduction, with COD decreasing from 10
856 ppm to 7113 ppm. HEMC, while showing a reduction, had the lowest percentage decrease at 9.11%, with COD changing from 6522 ppm to 5928 ppm. Finally, cet-HEC exhibited a 27.78% reduction, with COD decreasing from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 671 ppm to 7707 ppm.
671 ppm to 7707 ppm.
| CRM type | Initial COD (ppm) 0th day | Final COD (ppm) 60th day | % Reduction in COD |
|---|---|---|---|
| a HPC: hydroxypropyl cellulose; HPMC: hydroxypropyl methyl cellulose; HEMC hydroxyethyl methyl cellulose; cet-HEC: cetyl-hydroxyethyl cellulose. | |||
| HPC-J | 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 136 136 |
59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280 |
16.7 |
| HPC-M | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 640 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 784 784 |
40.0 |
| HPMC | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 856 856 |
7113 | 40.0 |
| HEMC | 6522 | 5928 | 9.11 |
| cet-HEC | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 671 671 |
7707 | 27.8 |
The observed reductions in COD values after 56 days of incubation indicate that some of the organic material from the CRMs was indeed consumed or mineralized during the biodegradation process. COD directly measures the amount of oxygen required to chemically oxidize organic and inorganic matter in a sample; thus, a decrease suggests a reduction in the total amount of oxidizable organic compounds present. However, the finding that the COD reduction (ranging from 9.11% to 40.01%) was not as significant as the substantial reductions observed in viscosity and molecular weight (which often indicated near-complete degradation of rheological properties and polymer chains) provides crucial insight into the degradation process. The dramatic reductions in viscosity and molecular weight point to rapid and extensive polymer chain hydrolysis (primary degradation), breaking down large macromolecules into much smaller fragments. A key justification for the less distinct COD reduction is that these intermediate breakdown products (e.g., oligomers and monomers like glucose) are still organic compounds. While they have lost their rheological properties and high molecular weight, they still possess a high chemical oxygen demand until they are fully mineralized into CO2, water, and biomass.
Comprehensively, the multi-metric approach proposed in this study effectively distinguishes between different phases of biodegradation and provides a robust framework for early-stage screening, which is critical for novel CRMs. The monitoring parameters can be categorized as follows:
(a) Primary degradation (chain scission): viscosity and molecular weight (Mw) reduction are highly sensitive, nearly instantaneous indicators of the microbial/enzymatic cleavage of the polymer backbone. The strong correlation between viscosity loss and polymer chain breakdown is well-established in polymer science,32 and our GPC data confirm this correlation (e.g., HPMC's near-100% viscosity loss coincided with a 46.1-fold Mw decrease). This metric identifies if a material is susceptible to initial breakdown.
(b) Substrate consumption (mass loss): total carbohydrate content (TCC) reduction serves as a direct, quantitative measure of the loss of the parent material from the system as it is broken down, consumed, and assimilated by the microbial consortia. TCC reduction generally correlated well with primary degradation metrics (e.g., HPC-J showed the highest TCC loss, 85%, and significant Mw loss, 3.0-fold), indicating effective breakdown and utilization of the resulting oligomers.
(c) Ultimate degradation (mineralization proxy): chemical oxygen demand (COD) reduction acts as a proxy for the total oxidation and potential mineralization of the organic compounds. The observed low COD reduction (9–40%) compared to the high viscosity/Mw loss highlights a critical finding; most of the CRMs rapidly undergo primary degradation but achieve only partial ultimate degradation within the 56-day period. This suggests the rapid accumulation of low-Mw, persistent intermediates that still retain oxidizable carbon (high COD), which would not be detected as ‘biodegradable’ by a standard, singular CO2 evolution test until much later.
By combining these four metrics, the developed method offers a comprehensive assessment; it identifies materials that are easily cleaved (HPMC, HPC-M, and cet-HEC via viscosity/Mw), quantifies the extent of mass loss (HPC-J via TCC), and most importantly, flags potential issues with slow final mineralization (low COD for all samples). This integrated approach provides a robust decision tool for early-stage CRM development.
3.5 Predictive analysis for polymer degradation (sensitivity factor)
Beyond the direct physiochemical and concentration analyses carried out for assessing the biodegradability of CRMs, a mathematical modelling approach was employed to derive a predictive degradation metric. Specifically, the linear regression model (S6, SI) was applied to the viscosity and GPC data across the 56-day test period to compute the degradation sensitivity factors (‘a’) and correlation coefficients (R2) for each CRM; these results are furnished in Table 3.| CRM type | Predictive exponent (a) | (R2) | p-Value | Std error | Interpretation |
|---|---|---|---|---|---|
| a HPC: hydroxypropyl cellulose; HPMC: hydroxypropyl methyl cellulose; HEMC hydroxyethyl methyl cellulose; cet-HEC: cetyl-hydroxyethyl cellulose. | |||||
| HPC-J | 1.42 | 0.90 | 0.014 | 0.28 | Strong correlation and statistically significant relationship; viscosity moderately tracks molecular weight loss |
| HPC-M | 2.19 | 0.99 | 0.0007 | 0.15 | Excellent correlation and viscosity is highly sensitive to degradation; clean power-law behavior |
| HPMC | 0.48 | 0.91 | 0.013 | 0.09 | Very stable polymer and viscosity retained despite scission; network reinforcement likely |
| HEMC | 0.93 | 0.78 | 0.047 | 0.28 | Moderate correlation; viscosity tracks molecular scission modestly |
| cet-HEC | 4.85 | 0.99 | 0.0003 | 0.25 | Extremely sensitive system; rapid viscosity collapse with small molecular weight loss |
The results show a wide range of ‘a’ values, confirming that the chemical structure significantly modulates the relationship between polymer chain scission (molecular weight loss) and functional loss (viscosity reduction). The highest ‘a’ value was observed for cet-HEC (a = 4.85), indicating that it is the most degradation-sensitive polymer; a small reduction in molecular weight results in a highly increased (nearly five-fold) loss of viscosity. This extreme sensitivity aligns with the rapid initial viscosity reduction observed for cet-HEC (Fig. 1). HPC-M (a = 2.19) also exhibited high sensitivity, which is within the range commonly cited for entangled polymer systems,33 showing an excellent fit (R2 = 0.99) and signifying a clean power-law behavior between molecular weight and viscosity loss.2
Conversely, HPMC displayed the lowest sensitivity factor (a = 0.48), yet demonstrated the highest goodness of fit (R2 = 0.91). This indicates that while its viscosity loss is perfectly predictable from its molecular weight reduction, the functional loss is significantly less sensitive to chain scission than in the other polymers. This low ‘a’ value strongly suggests that the structural modifications in HPMC provide maximum resistance to the chain scission process, requiring extensive breakdown before a significant functional property loss is measured.
This quantitative factor ‘a’, therefore, provides a predictive score for assessing structural resistance to biodegradation exhibited by the cellulosic-rheology modifiers, which can be determined early in the research & development phase.
4 Conclusions
We successfully established and validated a new multi-metric approach for early-stage biodegradability screening of cellulose-based rheology modifiers (CRMs). This method primarily utilized viscosity reduction as a key indicator, comprehensively supported by molecular weight, total carbohydrate content (TCC), and chemical oxygen demand (COD) analyses under ecologically relevant conditions. All five tested CRMs exhibited significant viscosity reduction, approaching 100% within 28–42 days for HPC-J, HPC-M, and cet-HEC, directly signifying polymer chain breakdown. Molecular weight analyses validated this, with HPMC demonstrating an impressive 46.1-fold reduction. The consistent and extensive reduction in TCC (up to ∼90% for HPC-J) provided robust quantitative evidence of microbial consumption and assimilation.Crucially, a quantitative modelling approach was integrated to establish a predictive mathematical relationship (degradation sensitivity factor, ‘a’) between viscosity loss and molecular degradation. The ‘a’ factor quantified the polymer's structural resistance, ranging from the most sensitive polymer, cet-HEC (a = 4.85), to the most structurally resistant, HPMC (a = 0.48). This factor enhances the predictive capability of the method by offering a metric for assessing structural stability during the R&D phase. The developed viscosity-based multi-metric method is simple, cost-effective, and rapid, making it highly suitable for preliminary screening and quality control. Its findings align with established biodegradability frameworks, holding significant promise for accelerating the development of sustainable rheology modifiers.
Author contributions
M. B. performed all the experiments, carried out analysis, data acquisition & prepared the primary draft. C. T. designed the experiments, interpreted the data, and wrote & edited the manuscript. A. R. supported in the setup of experiments & carried out GPC analysis. H. R. V. conceptualized the biodegradation tests, supervised the research, reviewed, edited, proofread and approved the manuscript. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare. This research was conducted by Alt Material (altM) Innovations Private Limited, a funded startup. The work was made possible through financial support from private investors and/or institutional funding. The authors serve as reviewers for other scientific journals, but this role has no impact on the work described in this manuscript.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: (S1) CRM chemical structures of HPC, HPMC, HEMC, and cet-HEC, (S2) chromatograms of GPC analysis (calibration curves), (S3) chromatograms of GPC analysis (test samples), (S4) total carbohydrate analysis (calibration curve), (S5) methodology for the synthesis of hydroxypropyl celluloses, (S6) composition of minimal media (stock concentrations), (S7) results of quantitative mathematical modeling, (S8) viscosity profiles of CRMs during the initial phase of biodegradation, and (S9) SDS-PAGE gel profile of proteins. See DOI: https://doi.org/10.1039/d5su00787a.Acknowledgements
The authors thank Alt Material Innovations Private Limited (short form altM), Bangalore for the financial support to carry out this research work. The authors acknowledge Mr Bharath Bakaraju (Founder and CEO of Phyx44 Private Limited, Bangalore) and Dr Jayesh Varavadekar for helping with the SDS-PAGE gel analysis.References
- K. Duis, T. Junker and A. Coors, Environmental fate and effects of water-soluble synthetic organic polymers used in cosmetic products, Environ. Sci. Eur., 2021, 33(1), 21 CrossRef CAS.
- P. G. Jessop, F. Ahmadpour, M. A. Buczynski, T. J. Burns, N. B. Green and R. Korwin, et al., Opportunities for greener alternatives in chemical formulations, Green Chem., 2015, 17(5), 2664–2678 RSC.
- L. Gandolfi and R. Galleguillos, Rheology modifiers and consumer perception, in Harry's Cosmeticology, ed. M. R. Rosen, Wiley, New York, 9th edn, 2015, pp. 768–806 Search PubMed.
- Y. J. Zheng and X. J. Loh, Natural rheological modifiers for personal care, Polym. Adv. Technol., 2016, 27(12), 1664–1679 CrossRef CAS.
- A. Samir, F. H. Ashour, A. A. Hakim and M. Bassyouni, Recent advances in biodegradable polymers for sustainable applications, npj Mater. Degrad., 2022, 6(1), 68 CrossRef CAS.
- N. R. Nair, V. C. Sekhar, K. M. Nampoothiri and A. Pandey, Biodegradation of biopolymers, in Current Developments in Biotechnology and Bioengineering: Production, Isolation and Purification of Industrial Products, Elsevier, Amsterdam, 2017, pp. 739–755 Search PubMed.
- N. T. Dintcheva, Overview of polymers and biopolymers degradation and stabilization towards sustainability and materials circularity, Polymer, 2024, 306, 127136 CrossRef CAS.
- V. C. Albright and Y. Chai, Knowledge gaps in polymer biodegradation research, Environ. Sci. Technol., 2021, 55(17), 11476–11488 CrossRef PubMed.
- A. M. El Mahdi and H. A. Aziz, A review on biodegradation and toxicity methods: risk assessment, standards, and analyses, in Toxicity and Biodegradation Testing, Taylor & Francis, London, 2017, pp. 349–388 Search PubMed.
- Organisation for Economic Co-operation and Development (OECD), Test No. 301: ready biodegradability, OECD Guidelines for the Testing of Chemicals, Section 3, OECD Publishing, Paris, 1992 Search PubMed.
- International Organization for Standardization (ISO), ISO 14851:2019, Determination of the ultimate aerobic biodegradability of plastic materials in an aqueous medium: method by measuring the oxygen demand in a closed respirometer, ISO, Geneva, 2019 Search PubMed.
- S. Takekoshi, K. Takano, Y. Matoba, M. Sato and A. Tachibana, Investigation of OECD 301F ready biodegradability test to evaluate chemical fate in a realistic environment, J. Pestic. Sci., 2021, 46(2), 143–151 CrossRef CAS.
- H. Loonen, F. Lindgren, B. Hansen, W. Karcher, J. Niemelä and K. Hiromatsu, et al., Prediction of biodegradability from chemical structure: modeling of ready biodegradation test data, Environ. Toxicol. Chem., 1999, 18(8), 1763–1768 CrossRef CAS.
- S. A. Miller, Biodegradable polymers: a macro perspective, ACS Macro Lett., 2013, 2(6), 550–554 CrossRef CAS.
- S. Ghosh, R. Chowdhury and P. Bhattacharya, Mixed consortia in bioprocesses: role of microbial interactions, Appl. Microbiol. Biotechnol., 2016, 100(10), 4283–4295 CrossRef CAS.
- M. DuBois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Colorimetric method for determination of sugars and related substances, Anal. Chem., 1956, 28(3), 350–356 CrossRef CAS.
- P. C. Hiemenz and T. P. Lodge, Polymer Chemistry, CRC Press, Boca Raton (FL), 2nd edn, 2007 Search PubMed.
- H. Lü, J. L. Wei, G. X. Tang, Y. S. Chen, Y. H. Huang and R. Hu, et al., Microbial consortium degrading of organic pollutants: source, degradation efficiency, pathway, mechanism and application, J. Clean. Prod., 2024, 451(3), 141913 CrossRef.
- N. Mohanan, Z. Montazer, P. K. Sharma and D. B. Levin, Microbial and enzymatic degradation of synthetic plastics, Front. Microbiol., 2020, 11, 580709 CrossRef PubMed.
- V. M. Pathak, Navneet. Review on the current status of polymer degradation: a microbial approach, Bioresour Bioprocess, 2017, 4(1), 1–31 CrossRef.
- A. Tayal, R. M. Kelly and S. A. Khan, Rheology and molecular weight changes during enzymatic degradation of a water-soluble polymer, Macromolecules, 1999, 32(2), 294–300 CrossRef CAS.
- International Organization for Standardization (ISO), ISO 17088:2021, Plastics: specifications for compostable plastics, ISO, Geneva, 3rd edn, 2021 Search PubMed.
- ASTM International, ASTM D5338-21, Standard test method for determining aerobic biodegradation of plastic materials under controlled composting conditions, incorporating thermophilic temperatures, ASTM International, West Conshohocken (PA), 2021 Search PubMed.
- R. Solaro, A. Corti and E. Chiellini, Biodegradation of poly(vinyl alcohol) with different molecular weights and degree of hydrolysis, Polym. Adv. Technol., 2000, 11(8–12), 873–878 CrossRef CAS.
- G. Robinson, S. B. Ross-Murphy and E. R. Morris, Viscosity-molecular weight relationships, intrinsic chain flexibility, and dynamic solution properties of guar galactomannan, Carbohydr. Res., 1982, 107(1), 17–32 CrossRef CAS.
- B. Saake, S. Horner, T. Kruse, J. Puls, T. Liebert and T. Heinze, Detailed investigation on the molecular structure of carboxymethyl cellulose with unusual substitution pattern by means of an enzyme-supported analysis, Macromol. Chem. Phys., 2000, 201(15), 1996–2002 CrossRef CAS.
- Y. Diao, M. Song, Y. Zhang, L. Y. Shi, Y. Lv and R. Ran, Enzymic degradation of hydroxyethyl cellulose and analysis of the substitution pattern along the polysaccharide chain, Carbohydr. Polym., 2017, 169, 92–100 CrossRef CAS.
- H. Schagerlöf, S. Richardson, D. Momcilovic, G. Brinkmalm, B. Wittgren and F. Tjerneld, Characterization of chemical substitution of hydroxypropyl cellulose using enzymatic degradation, Biomacromolecules, 2006, 7(1), 80–85 CrossRef.
- M. Nojiri and T. Kondo, Application of regioselectively substituted methylcelluloses to characterize the reaction mechanism of cellulase, Macromolecules, 1996, 29(7), 2392–2395 CrossRef CAS.
- H. Seddiqi, E. Oliaei and H. Honarkar, et al., Cellulose and its derivatives: towards biomedical applications, Cellulose, 2021, 28(3), 1893–1931 CrossRef CAS.
- L. R. Lynd, P. J. Weimer, W. H. Van Zyl and I. S. Pretorius, Microbial cellulose utilization: fundamentals and biotechnology, Microbiol. Mol. Biol. Rev., 2002, 66(3), 506–577 CrossRef CAS.
- G. Robinson, S. B. Ross-Murphy and E. R. Morris, Viscosity-molecular weight relationships, intrinsic chain flexibility, and dynamic solution properties of guar galactomannan, Carbohydr. Res., 1982, 107(1), 17–32 CrossRef CAS.
- B. E. Dolata, S. L. Rosen and S. T. Milner, Entanglement kinetics in polymer melts are chemically universal, Nat. Commun., 2024, 15, 12345 Search PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |