Open Access Article
Open Access ArticleFluorogenic squaraine dimers for the flow cytometry detection of bacterial urinary tract infections
Lucille Weissa,
Martijn Visser a,
Patrick Wagnera,
Anthony Augéa,
Océane Florèsa,
Nicola Pordone
a,
Patrick Wagnera,
Anthony Augéa,
Océane Florèsa,
Nicola Pordone bc,
Martin Fayolle
bc,
Martin Fayolle bcd,
Paul O. Verhoeven
bcd,
Paul O. Verhoeven bcd,
Dmytro Dziuba
bcd,
Dmytro Dziuba e,
Dominique Bonnet
e,
Dominique Bonnet a and
Julie Karpenko
a and
Julie Karpenko *af
*af
aLaboratoire d'Innovation Thérapeutique, UMR7200 CNRS/Université de Strasbourg, Institut du Médicament de Strasbourg, F-67000 Strasbourg, France. E-mail: julie.karpenko@unistra.fr
bCIRI, Centre International de Recherche en Infectiologie, Université de Lyon, Université Jean Monnet St-Etienne, Inserm, U1111, CNRS, UMR5308, ENS Lyon, Université Claude Bernard Lyon 1, Lyon, France
cFaculté de Médecine, Université Jean Monnet St-Etienne, St-Etienne, France
dService des Agents Infectieux et Hygiène, Centre Hospitalier Universitaire de St-Etienne, St-Etienne, France
eLaboratoire de Bioimagerie et Pathologies, UMR7021 CNRS/Université de Strasbourg, F-67000 Strasbourg, France
fInstitut Universitaire de France (IUF), France
First published on 28th November 2025
Abstract
Rapid diagnosis of bacterial infections is essential for guiding antibiotic treatment and mitigating the global spread of antibiotic resistance. Here, we report the development of fluorogenic bacteria-specific probes for rapid flow cytometry detection of urinary tract infections. These probes, composed of a covalently linked pair of squaraine dyes conjugated to a bacteria-specific peptidic vector, exploit the phenomenon known as aggregation-caused quenching. The chemical structure of the squaraine dyes was finely tuned to ensure selectivity for bacteria over eukaryotic cells and human serum albumin. The most effective probe enabled semi-quantitative detection of bacteria in patient urine samples, with a total time-to-result of less than one hour.
Introduction
Due to the planetary spread of antibiotic resistance, bacterial infections have become one of the most critical dangers facing modern society.1–3 The number of deaths from multidrug-resistant bacteria is rising and, without new antimicrobial therapeutics, could reach 10 million deaths annually by 2050.4 Although the development of new antibacterial therapeutics and vaccines is of high priority,5 additional measures should be taken to develop rapid methods for the early diagnosis of bacterial infections to reduce the misuse and the overuse of antibiotics.The standard clinical laboratory methods for diagnosing urinary tract infections (UTI) rely on culturing bacteria from patient samples on semi-solid growth media to confirm a UTI, followed by identification of the pathogen and its susceptibility by MALDI-TOF, PCR, and other analytical methods.6 Although urine culture is still considered the “gold standard” in UTI diagnosis, it is typically time-consuming (24–48 hours), labor-intensive, and prone to false-positive results. Moreover, the majority of urine samples analyzed in the microbiology laboratories are negative for pathogens.7 An ideal next-generation diagnostic approach should begin with a rapid screening method to exclude UTI-negative urine samples, followed by a molecular technique for pathogen identification in UTI-positive samples, and, ultimately, determination of its antibiotic susceptibility. Such a diagnostic approach would not only shorten the diagnosis delay but also reduce the burden on biomedical laboratories by cutting back on the number of plates to analyze to those with defined UTI.
Significant efforts have been made to develop rapid and direct screening methods for the detection of UTIs without the need for urine culture, using techniques such as lateral flow immunoassays, flow cytometry, PCR, FISH, and diagnostics platforms based on biosensors or microfluidics.7,8 Among these methods, urine flow cytometry appears promising, as it enables direct, rapid, and inexpensive prediction of pathogen-positive urine samples for further analysis.9–12 However, its diagnostic accuracy requires improvements before the method can be widely adopted in clinical practice. One area where such improvement could be achieved through chemical approaches is the development of specific fluorescent bacteria-targeting probes compatible with urine flow cytometry.
Fluorescence holds great potential as a diagnostic modality due to its fast response, high sensitivity, and the broad range of available instrumentation, including inexpensive portable fluorescence readers.13–16 However, the design of bacteria-specific fluorescent probes that could operate in body fluids containing a wide range of biomolecules, metabolites, and human cells is a formidable challenge.17 Current flow cytometry-based screening for the presence of bacteria in urine either relies solely on light scattering or employs non-specific DNA-binding dyes, such as SYBR Green, which are not selective for bacterial cells and also stain human cells and cellular debris.11,18
An ideal bacteria-targeting probe for urine analysis should absorb and emit fluorescence in the red or far-red spectral region to avoid interference with the sample's autofluorescence, should be fluorogenic (become fluorescent only in the presence of bacteria and remain dark in their absence), and possess high selectivity for bacteria against human cells and other constituents of urine (e.g. proteins, peptides, nucleic acids, metabolites, etc.). Several approaches have been explored to achieve fluorogenicity (or fluorescence turn-on) in the presence of bacteria. Fluorogenic bacteria-targeting probes have been conceived using environmentally sensitive organic fluorophores, such as push–pull dyes,19–22 molecular rotors,23,24 or rhodamine derivatives that undergo a spirolactone/lactam isomerization.25 In such a case, binding of the probe to bacteria produces changes in the microenvironment of the fluorophore, which leads to a fluorescence enhancement. Fluorescence turn-on has also been achieved upon aggregation of the probes operating by aggregation-induced emission (AIE)26 on the bacterial envelope, and upon activation of a pro-fluorescent probe by a bacterial enzyme.27
An alternative strategy involves using aggregation-caused quenching (ACQ) of aromatic dyes as an approach to reduce off-target fluorescence and increase the fluorogenicity of the probe.28 An ACQ-based probe is typically conceived in the form of a covalent dimer of lipophilic organic dyes linked by a flexible linker. In an aqueous environment, the hydrophobic interaction between two dyes leads to the formation of a non-fluorescent intramolecular H-aggregate. The H-aggregate is disturbed in a less polar environment, resulting in the fluorescence turn-on. This strategy has recently regained attention for the design of fluorogenic probes targeting biomolecules. We and others have applied the ACQ approach to develop fluorogenic probes for nucleic acids,29,30 membrane receptors,31–35 membrane lipids,36 enzymes,37 and cancer biomarkers,38 among others. It has been shown that a wide variety of organic dyes are prone to the formation of intramolecular H-aggregates in aqueous solutions. Examples are carbocyanine,32 squaraine,31 BODIPY,35 and rhodamine29 derivatives. The versatility of the ACQ approach enables the rational design of fluorogenic probes having a desired combination of properties, such as excitation and emission wavelengths, magnitude of fluorogenic response, photostability, and brightness.
In this work, we applied the ACQ strategy to design fluorogenic dimeric probes for bacteria, with the ultimate goal being the development of a highly specific probe for the rapid detection of bacteria in clinical urine samples.
Results and discussion
Design of fluorogenic dimeric probes targeting bacteria
We have previously reported fluorogenic ACQ-based probes for G protein-coupled receptors.31–33 Composed of covalent dimers of far-red aromatic dyes linked to a GPCR peptide ligand, the probes adopted a closed non-fluorescent conformation in an aqueous medium. Upon binding to the target, the dimer dissociated due to the interactions with the lipids in the vicinity of the receptor, leading to the fluorescence turn-on. We speculated that a similar principle could be used to design fluorogenic dimeric probes targeting bacterial cells. Such probes should include a bacteria-targeting vector to ensure strong binding to bacteria and selectivity against eukaryotic cells, conjugated by a flexible linker to a dye dimer (Fig. 1A). The interaction of the vector with the bacterial membranes should bring the dye dimer closer to the membrane. If the dyes themselves are prone to interact with the bacterial membranes, the dimer will open, leading to a fluorescence turn-on response.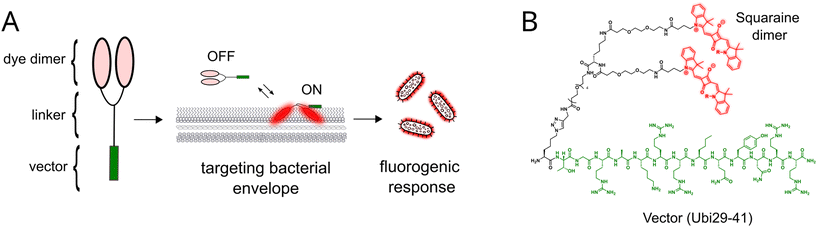 | ||
| Fig. 1 Functioning principle (A) and general structure (B) of the fluorogenic dimeric probes for bacteria described in this work. | ||
In such a design, the bacteria-targeting vector plays a crucial role in ensuring strong interactions with bacterial membranes and selectivity versus the membranes of eukaryotic cells. The vector should be easily synthesized on a semi-preparative scale, allowing a divergent synthesis of multiple dye dimers. The covalent conjugation of the vector to the dye dimer should not perturb the interactions of the former with the bacterial cell. Finally, the vector should enhance the aqueous solubility of the conjugate to counterbalance the hydrophobic nature of the fluorophores, since we have recently demonstrated that the best fluorescence turn-ons are achieved using the dimers of hydrophobic water-insoluble dyes.33 Taking into account the criteria mentioned above, we decided to use the antimicrobial peptide Ubi29-41 as the vector. Ubi29-41 is a linear positively charged thirteen amino acid-long peptide that non-covalently binds to a wide range of Gram-positive and Gram-negative bacteria.39,40 This binding presumably relies on electrostatic interactions between the positively charged residues of Ubi29-41 and the negatively charged bacterial membranes. Due to its versatility, straightforward chemical synthesis, and amenability to modifications with fluorophores, Ubi29-41 has been used for constructing bacteria-targeting molecular imaging probes for in vitro detection and in vivo imaging.20,21,41–44
We designed our bacteria-targeting fluorogenic probes based on Ubi29-41 with the methionine to norleucine substitution at position 36 to increase their stability toward oxidation.21 An additional amino acid, azidolysine Lys(N3), was added to the N-terminus of the peptide to enable conjugation of dye dimers using CuAAC (copper-catalyzed azide-alkyne cycloaddition) click chemistry (Fig. 1B). As for the dye dimers, their design relied on a branched PEG linker between the dyes and the vector. An L-lysine residue was used to create the branching point in the linker. A short spacer of two PEG units was introduced between the fluorophores and the branching point to ensure the conformational flexibility of the fluorophore pair, and a PEG4 unit was inserted to distance the vector from the branching point. Squaraine fluorophores were chosen as a reporter unit of the probe due to their high brightness, far-red excitation and emission, high chemical and photostability, and a rather straightforward modular synthesis allowing for tuning their physico-chemical and biochemical properties.31,45 The general structure of the fluorogenic bacteria-targeting dimeric probes developed in this work is shown in Fig. 1B.
Addition of lipidic or positively charged substituents increased the capacity of squaraines to label bacteria
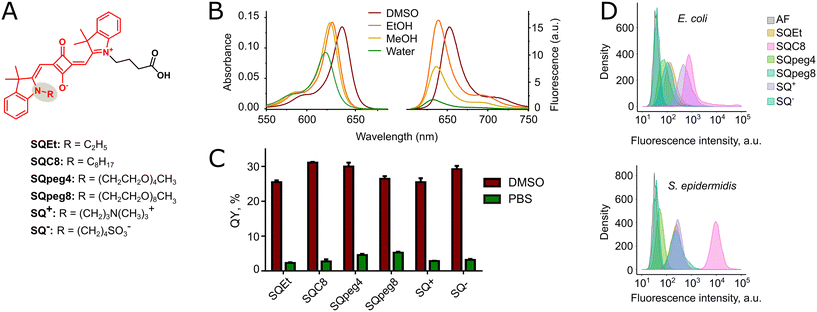
To ensure the correct transition of the fluorogenic dimer from the closed quenched to the opened fluorescent conformation upon binding to the bacterial cell, squaraine dyes constituting the dimer should have an intrinsic affinity for the bacterial cell. Therefore, we designed and synthesized a small set of asymmetric squaraines to determine how the presence of apolar, polar, and charged substituents influenced the photophysical properties of the fluorophores and their interaction with bacteria (Fig. 2A). The fluorophores were synthesized using a condensation reaction between squaric acid and 2,3,3-trimethylindolenines N-alkylated with different substituents: ethyl (SQEt), octyl (SQC8), PEG4 (SQpeg4), PEG8 (SQpeg8), trimethylpropylammonium (SQ+), and butylsulfonate (SQ−). SQEt and SQpeg4 were synthesized following the previously published protocols.31 The details of the synthesis of squaraines SQC8, SQpeg8, SQ+, and SQ− are presented in the SI.All the squaraines absorbed light in the red spectral region, displaying molar absorption coefficients in methanol in the range 221![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–278
000–278![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 (Table 1). When excited at 600 nm, the squaraines displayed a sharp fluorescence emission band with a Stokes shift of about 10 nm (Fig. 2B and Table 1). A negative solvatochromism could be deduced from the analysis of absorption and emission spectra, especially when analyzing polar protic solvents. It manifests as a blue shift of the absorption and emission spectra upon increased solvent polarity. Thus, the absorption maxima for SQEt in EtOH, MeOH, and water were 632 nm, 630 nm, and 623 nm, respectively. The fluorescence emission maxima followed the same tendency: for EtOH, MeOH, and water, they were 639 nm, 636 nm, and 633 nm, respectively.
000 M−1 cm−1 (Table 1). When excited at 600 nm, the squaraines displayed a sharp fluorescence emission band with a Stokes shift of about 10 nm (Fig. 2B and Table 1). A negative solvatochromism could be deduced from the analysis of absorption and emission spectra, especially when analyzing polar protic solvents. It manifests as a blue shift of the absorption and emission spectra upon increased solvent polarity. Thus, the absorption maxima for SQEt in EtOH, MeOH, and water were 632 nm, 630 nm, and 623 nm, respectively. The fluorescence emission maxima followed the same tendency: for EtOH, MeOH, and water, they were 639 nm, 636 nm, and 633 nm, respectively.
| Fluorophore | λabs,a nm | λfl,b nm | ε,c M−1 cm−1 | QYd | Turn-ong | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| DMSO | EtOH | MeOH | Water | PBSe | LBf | |||||
| a Position of the absorption maximum in MeOH.b Position of the emission maximum in MeOH.c Molar absorption coefficient determined at the absorption maximum in MeOH.d Fluorescence quantum yields expressed as means from three independent experiments.e Phosphate-buffered saline.f Luria-Bertani broth.g Calculated as the ratio between the fluorescence quantum yields in DMSO and PBS. | ||||||||||
| SQEt | 630 | 636 | 267![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.25 | 0.19 | 0.08 | 0.02 | 0.02 | 0.03 | 13 |
| SQC8 | 630 | 637 | 221![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.31 | 0.22 | 0.11 | 0.01 | 0.03 | 0.03 | 10 |
| SQpeg4 | 630 | 638 | 278![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.30 | 0.26 | 0.11 | 0.06 | 0.04 | 0.05 | 8 |
| SQpeg8 | 630 | 639 | 264![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.26 | 0.21 | 0.11 | 0.05 | 0.05 | 0.06 | 5 |
| SQ+ | 630 | 639 | 229![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.25 | 0.18 | 0.09 | 0.03 | 0.03 | 0.04 | 8 |
| SQ− | 629 | 638 | 241![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.29 | 0.25 | 0.11 | 0.03 | 0.03 | 0.04 | 10 |
It is worth mentioning that squaraines could be considered fluorogenic dyes, although in our view, this property is largely underexplored.46 The quantum yields of fluorescence, being quite low in aqueous media (<10%), rose to above 25% in less polar solvents such as DMSO (Fig. 2C). In this work, we compared the fluorogenicity of the dyes by calculating their fluorescence turn-on as the ratio between the fluorescence quantum yields in DMSO and phosphate-buffered saline (PBS) (Table 1). The previously reported squaraine dyes SQEt and SQpeg4 displayed a modest magnitude of the fluorescence turn-on response (13- and 8-fold, respectively). The calculated fluorescence turn-on ratio for newly synthesized squaraines SQC8, SQpeg8, SQ+, and SQ− fell between 5- and 10-fold.
Next, we examined whether some of the synthesized squaraines could label bacteria per se, without conjugation to a bacteria-targeting vector. We used Escherichia coli and Staphylococcus epidermidis as model Gram-negative and Gram-positive bacteria, respectively. Bacteria in the early logarithmic growth phase (OD600 ≈ 0.5) were directly labeled in LB (Luria-Bertani broth, a nutrient-rich growth medium) with 1 μM squaraines for 10 min, then fixed with paraformaldehyde (4% w/v) and analyzed by flow cytometry. The results are presented in Fig. 2D and S1. For the Gram-negative bacterium E. coli, the brightest labeling was achieved using SQC8 and SQ+, followed by SQEt. In the case of the Gram-positive bacterium S. epidermidis, SQC8 was found to be much more efficient compared to the other dyes. Moreover, SQC8 labeled Gram-positive bacteria with intensities almost ten times higher than Gram-negative bacteria. On the other hand, SQ+, SQEt, and SQ− were also capable of labeling S. epidermidis with intensities comparable to those obtained for the Gram-negative bacteria. To summarize, SQC8 was found to be the most efficient squaraine to label both Gram-negative and Gram-positive bacteria, presumably due to hydrophobic interactions of the C8 substituent with lipidic constituents of the bacterial membranes. SQC8 was closely followed by SQ+, bearing a positively charged substituent, which should electrostatically interact with the negatively charged components of bacterial cell envelopes (e.g. lipopolysaccharides in Gram-negative bacteria or teichoic acids in Gram-positive bacteria). Next, SQEt, deprived of lipophilic or positively charged substituents, demonstrated modest bacterial labeling. Finally, the less lipophilic dyes SQpeg4, SQpeg8, and the negatively charged SQ− were the least efficient in labeling both Gram-negative and Gram-positive bacteria.
Based on these results, we decided to synthesize dimeric probes targeting bacteria from the three most efficient squaraines: SQC8, SQ+, and SQEt.
Synthesis of dimeric conjugates targeting bacteria
The key step of the synthesis of dimeric probes for bacteria is the CuAAC click reaction between the antimicrobial peptide Ubi29-41 modified with an azide group (LysN3-Ubi), and the squaraine dimer bearing a terminal alkyne group (Scheme 1).We synthesized LysN3-Ubi on the solid phase following the classical Fmoc/tBu approach. Lys(N3) was added at the N-terminus of the peptide to enable the CuAAC conjugation. The peptide was obtained by automatic microwave-assisted synthesis with a 36% isolation yield and an LC-HRMS purity of >95%.
Diamino-alkyne PEG linker 1 was synthesized in solution, starting from commercially available Boc-protected amino-PEG-acids and Boc-L-Lys(Boc)-OH. The details of the synthesis are presented in the SI.
SQEt, SQC8, and SQ+ were coupled to 1 under PyBOP in situ activation in the presence of DIEA in DMF, to give dimers 2, 3, and 4 (Scheme 1). The CuAAC conjugation of dimers 2 and 4 to LysN3-Ubi was performed in the presence of copper(II) sulfate and sodium ascorbate in a mixture of DMSO and water to obtain dSQEt-Ubi and dSQ+-Ubi in satisfactory yields. As for dimer 3, its low water solubility did not allow us to use even a low percentage of water in the reaction mixture, required to solubilize the salts. We thus used a DMSO-soluble catalyst, [(CH3CN)4Cu]PF6, which afforded the desired conjugate dSQC8-Ubi in 22% yield after purification.
In parallel, the monomeric conjugate mSQC8-Ubi, bearing only one fluorophore, was prepared by grafting the most efficient squaraine SQC8 to the peptide vector LysN3-Ubi via a linear PEG8 chain. The synthesis of this monomeric conjugate, which is essential for assessing the impact of dimerization on photophysical properties and bacterial specificity, is provided in the SI.
Dimerization of squaraines increased the fluorescence turn-on between organic and aqueous solvents
First, the impact of the dimerization of squaraines on the spectroscopic properties was evaluated. The absorption spectra of all the dimeric conjugates in aqueous solutions possessed a short-wavelength maximum with a long-wavelength shoulder, which is characteristic of the formation of non-fluorescent intramolecular H-aggregates (Fig. 3A and S2).31 These aggregates were distorted in organic solvents, giving absorption spectra similar to those of the non-dimerized squaraine dyes. Conversely, the monomeric conjugate mSQC8-Ubi was characterized by the absorption spectra similar to those of the non-conjugated squaraine dyes without any visible sign of aggregation (Fig. S2).The fluorescence quantum yields of the dimeric conjugates in aqueous solutions decreased compared to squaraine dyes, which confirmed that these conjugates exist preferentially in the closed, non-fluorescent form in aqueous media (Table S1). In organic solvents such as DMSO, the fluorescence quantum yields were similar to those of the non-dimerized squaraines, suggesting the efficient opening of the dimers when the fluorophores are solvated. The ability of the dimers to open and close, depending on the solvent, led to a strong fluorescence turn-on between the organic and aqueous media, expressed as a ratio between the quantum yields in DMSO and PBS. For the dimeric conjugates, the fluorescence turn-on ratios ranged between 31 and 42, while for the corresponding non-dimerized dyes these values fell between 8 and 13 (Table 1). The monomeric conjugate mSQC8-Ubi exhibited significant fluorescence in aqueous media with fluorescence quantum yields of 10%, resulting in a modest turn-on ratio of 3 (Table S1).
Dimeric conjugates efficiently labeled model Gram-positive and Gram-negative bacteria
Next, we examined the capacity of the fluorescent conjugates dSQEt-Ubi, dSQC8-Ubi, dSQ+-Ubi, and mSQC8-Ubi to label model Gram-negative and Gram-positive bacteria using the same protocol as for the evaluation of squaraine dyes. In brief, E. coli and S. epidermidis in the early logarithmic growth phase were directly stained in LB with 1 μM conjugates for 10 min, then fixed with 4% PFA and analyzed by flow cytometry.All the conjugates efficiently labeled model bacteria, regardless of their dimeric or monomeric nature (Fig. 3B and S1). Moreover, comparison of the fluorescence intensities obtained for non-conjugated squaraines and the Ubi29-41 conjugates revealed that all the conjugates were more efficient in bacterial labeling than the non-conjugated squaraine dyes, except for SQC8 derivatives. This finding underscores the critical role of the bacteria-targeting vector (Ubi29-41), which enhanced bacterial labeling even with the dyes having the lowest affinity to the bacterial envelope. Among the dimeric conjugates, the positively charged squaraine derivative dSQ+-Ubi provided the most effective labeling of both E. coli and S. epidermidis.
Dimerization of squaraines decreased off-target fluorescence in the presence of human serum albumin and eukaryotic cells
Next, we aimed to evaluate if the dimeric conjugates were prone to fluoresce in the presence of serum proteins and eukaryotic cells. Such off-target fluorescence could dramatically decrease the signal-to-noise ratio of the bacterial turn-on detection. UTIs are often associated with pyuria, which refers to the presence of high levels of leukocytes in the urine.47 To be used directly in urine samples, the fluorescent probe for UTI should be characterized by a high selectivity for bacteria against eukaryotes. On the other hand, urine contains small amounts of albumin, and its level increases during albuminuria. Although the association of UTI with albuminuria is not very common, one should not exclude such a possibility.We used a solution of 0.1% HSA (human serum albumin) in PBS to estimate the undesired fluorescence turn-on in the presence of urine albumins (Fig. 3C). The non-conjugated squaraine dyes SQEt, SQC8, and SQ+ were characterized by a strong fluorescence in the presence of HSA, with quantum yields reaching 75% in case of SQC8. One possible explanation for the strong fluorescence turn-on in the presence of HSA could be the binding of the non-conjugated dyes to one of the hydrophobic binding pockets of the protein, which leads to a restriction of their intramolecular rotation. This restriction suppresses non-radiative decay pathway consequently increases the fluorescence quantum yields, which is a characteristic behavior of molecular rotors. This observation is consistent with recent reports that have proposed the use of squaraine dyes as fluorogenic viscosity probes based on their molecular rotor properties.48,49 Similar fluorescence turn-on in the presence of HSA was observed for the monomeric conjugate dSQC8-Ubi. Despite its conjugation to the water-soluble Ubi29-41 vector via a PEG8 linker, the squaraine dye retained a high affinity for HSA, resulting in a fluorescence quantum yield of 61% in the presence of 0.1% HSA.
In contrast, dimerization of squaraines largely diminished their fluorescence in the presence of HSA (Fig. 3C). We hypothesized that the decreased fluorescence of the dimers in the presence of HSA could be attributed to the formation of strong intramolecular aggregates, which were not distorted upon interaction with the hydrophobic pocket of HSA. Among all the squaraine derivatives, SQ+ and its dimer demonstrated the lowest fluorescence in the presence of HSA (quantum yields of 11% and 3%, respectively). To determine whether the positive charge on the squaraine dye plays a role in reducing its binding to HSA, we measured the fluorescence quantum yield of the negatively charged dye SQ− in the presence of HSA. In contrast to SQC8 and SQEt, both SQ+ and SQ− are more water-soluble, have similar molecular size, and differ only by the opposite charge of the substituent on the squaraine core. In the presence of 0.1% of HSA SQ− demonstrated a strong fluorescence with the quantum yield of 68%, compared to the modest quantum yield of 11% for SQ+. This data revealed that the positive charge on the squaraine core played a more important role in decreasing the non-specific interactions with HSA than the overall water solubility of the dye.
Next, the potential off-target fluorescence caused by binding to eukaryotic cells was evaluated. Freshly detached HEK293T cells were incubated with 1 μM of squaraines and their conjugates in DMEM/F-12 culture medium without phenol red for 10 min and directly analyzed by flow cytometry (Fig. 3D). The experiment revealed that all non-conjugated squaraines displayed higher labeling of HEK293T cells than the conjugates. This reduction of non-specific fluorescence could be attributed to the decreased interactions of the conjugates with the eukaryotic membranes due to the presence of the positively charged hydrophilic Ubi29-41 peptide vector. The labeling of the eukaryotic cells was the strongest in the case of SQC8, probably due to interactions of its octyl chain with the lipid membranes. Its monomeric and dimeric conjugates presented slightly lower, but still significant HEK293T cell staining. Among the squaraine dimers, dSQ+-Ubi presented the lowest staining of HEK293T cells, followed by dSQEt-Ubi.
A confocal microscopy experiment was then performed to take a closer look at the interaction of the dyes and their dimers with HEK293T cells. Live adherent HEK293T cells were incubated with 1 μM solutions of squaraine dyes or their conjugates in DMEM/F-12 culture medium without phenol red for 10 min, then imaged under the microscope. The results confirmed the strong binding of SQC8 to eukaryotic cells (Fig. 3E). However, the staining was not limited to the cell membrane. The dye also labeled intracellular structures, presumably internal lipid membranes. Analyzed by microscopy, staining of cells with the Ubi29-41 conjugates seemed negligible compared to SQC8. However, a closer analysis of images by adjusting the brightness and contrast parameters revealed that cellular staining could also be observed in the case of dSQC8-Ubi, dSQEt-Ubi, and mSQC8-Ubi (Fig. S3). Only dSQ+-Ubi was fully deprived of the ability to stain HEK293T cells, even using a stronger laser power, which was consistent with the flow cytometry results. Fluorescence microscopy also confirmed the ability of dSQ+-Ubi to stain bacteria at the cellular level (Fig. 3F).
In conclusion, dimerization of the new squaraine dyes and their conjugation to the Ubi29-41 peptide vector led to the development of fluorogenic bacteria-targeting probes. Dimerization reduced fluorescence in aqueous media, thereby enabling a strong fluorescence turn-on in less polar environments. Moreover, it was shown that dimerization is crucial for minimizing off-target fluorescence in the presence of serum albumin. On the other hand, conjugation to the positively charged peptide vector Ubi29-41 enhanced both the efficiency of bacterial staining and its specificity over eukaryotic cells.
The comparison of the dimeric conjugates derived from three best bacteria-staining squaraine dyes revealed that dSQ+-Ubi was characterized by the lowest off-target fluorescence both in the presence of HSA and eukaryotic cells. As mentioned above, the specificity of the probe is crucial for its direct application in urine patient samples without pre-enrichment steps. It was thus decided to select dSQ+-Ubi for further development as a fluorescent probe for UTI.
dSQ+-Ubi labeled Gram-positive and Gram-negative bacteria isolated from clinical urine samples
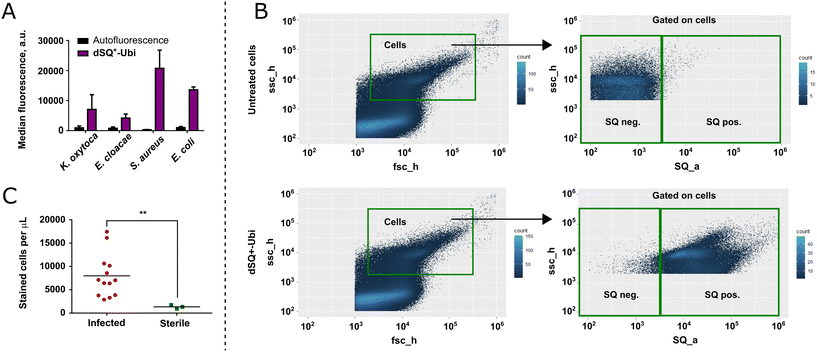
An important step before evaluating the performance of dSQ+-Ubi in patient urine samples was the assessment of the capacity of the probe to label clinical bacterial isolates responsible for UTI. Clinical isolates of Klebsiella oxytoca, Enterobacter cloacae, E. coli (Gram-negative bacteria), and Staphylococcus aureus (Gram-positive bacterium) were inoculated onto Columbia blood agar plates and incubated overnight at 37 °C. Three colonies of each isolated strain were resuspended in PBS to reach an OD600 of 0.5, labeled with 1 μM of dSQ+-Ubi for 10 min at room temperature, diluted 100 times, and analyzed by a commercial flow cytometer. The results presented in Fig. 4A demonstrated efficient labeling of all clinical isolates, with the highest fluorescence signal obtained for S. aureus.Towards the detection of UTI with fluorogenic dimers
Finally, the performance of dSQ+-Ubi in human urine samples was evaluated. A total of 16 urine samples collected into BD Vacutainer Urine Culture & Sensitivity tubes from patients with suspected UTI at the Saint-Etienne University Hospital Centre (France) were included in the study. After measuring cytological parameters (such as hematuria and leucocyturia), 10 μL of each sample were inoculated onto chromogenic UTI Clarity™ agar plates and incubated at 37 °C for 24 hours. The rest of the samples were kept overnight at +4 °C for further analysis using dSQ+-Ubi. Thirteen bacterial cultures yielded visible colonies, whereas the three remaining cultures showed no detectable growth after 24 hours of incubation. The identification of the bacterial species was performed by MALDI-TOF. The identified strains were Proteus mirabilis, E. coli, Klebsiella pneumoniae, Citrobacter koseri, Enterococcus faecium, Enterococcus faecalis, Streptococcus gallolyticus, and Staphylococcus saprophyticus. Two urine samples were co-infected with two organisms: K. pneumoniae and E. coli, E. coli and S. gallolyticus. The complete characterization of the collected urine samples is provided in Table S2.Once the pathogens in the urine samples were identified, we proceeded to a blinded analysis of the samples using dSQ+-Ubi. 100 μL of each urine sample was stained with dSQ+-Ubi at 1 μM concentration for 10 min at room temperature and fluorescence measurements were performed using a commercial flow cytometer. Exactly 5 μL of liquid were analyzed from each sample to allow for comparison of bacterial load across the samples. In parallel, untreated samples were analyzed in an identical manner. The instrumental settings of the cytometer were kept identical for both series of measurements. Overall, the analysis of each sample from labeling to the readout took less than 20 minutes.
The workflow of the data analysis is presented in Fig. 4B. It is essential to point out that bacteria are naturally present in urine of healthy individuals, but the bacterial burden is increased during UTIs (typically >103 CFU mL−1). Thus, the qualitative detection of stained bacteria in patient samples does not necessarily indicate a UTI. Rather, it is the difference in bacterial counts between sterile and infected urine samples that suggests a UTI.
The data analysis started with an initial gating by FSC/SSC to separate bacteria from cellular debris. Importantly, we used a large gate that should allow taking into account different bacterial strains, which could vary in size, shape and granularity. It is worth mentioning that such gating allows separating bacteria from cellular debris only partially, and for several samples, the bacterial population was not clearly visible in an FSC/SSC density plot due to an intense signal from debris. In such cases, it is particularly important to use a highly specific fluorescent probe, able to stain bacteria in a complex mixture. Next, the second gate in an SQ channel (red fluorescence 670–688 nm) versus SSC density plot was performed to separate stained and unstained cells. Finally, we calculated the number of stained cells per microliter for each sample, which should be proportional to the overall bacterial load of the sample.
The results of the analysis are presented in Fig. 4C. A strong fluorescence signal distinguishable from cellular autofluorescence was obtained in the gated population in all analyzed samples. As expected, a difference was observed between the number of detected stained cells in infected and sterile samples, regardless of the quantities of red and white blood cells (Table S2). As an example, the three sterile samples were characterized by the number of fluorescent events per microliter <2000. Among these samples, one was characterized by leucocyturia (23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 leucocytes per mL) and another by hematuria (11
000 leucocytes per mL) and another by hematuria (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 red blood cells per mL). In both cases, the number of fluorescent events per microliter stayed low. This confirmed the excellent specificity of the developed probe dSQ+-Ubi for bacteria, its universal labeling of both Gram-positive and Gram-negative bacteria, and high potential for further development of flow cytometry with dSQ+-Ubi for the rapid detection of UTIs.
000 red blood cells per mL). In both cases, the number of fluorescent events per microliter stayed low. This confirmed the excellent specificity of the developed probe dSQ+-Ubi for bacteria, its universal labeling of both Gram-positive and Gram-negative bacteria, and high potential for further development of flow cytometry with dSQ+-Ubi for the rapid detection of UTIs.
Conclusion
This work aimed to develop a specific fluorogenic bacteria-targeting probe for the flow cytometry detection of urinary tract infections. Chemical modification of the squaraine core led to the development of new far-red dyes with increased affinity for both Gram-positive and Gram-negative bacteria. The fluorogenic probes were synthesized by dimerizing the squaraine dyes via a flexible branched PEG linker and conjugating them to the peptidic bacteria-targeting vector Ubi29-41. We demonstrated that dimerization was essential for enhancing the probe's fluorogenicity and minimizing off-target fluorescence in the presence of serum albumin, while conjugation to the peptide vector ensured both efficient bacterial staining and high specificity over eukaryotic cells. The best probe, dSQ+-Ubi, exhibited an excellent fluorescence turn-on response and efficiently labeled both laboratory and clinically isolated bacterial strains with strong selectivity over eukaryotic cells and HSA. This probe enabled rapid and quantitative detection of bacteria in UTI samples by flow cytometry within minutes.Although our primary interest in dSQ+-Ubi lies in its application for urine flow cytometry, the probe may also find utility beyond the diagnostic domain. For example, it could be particularly suitable for confocal microscopy imaging of bacteria under no-wash conditions (Fig. 3F). Its fluorogenic properties and the lack of off-target fluorescence in the presence of albumins make it suitable for bacterial labeling in co-culture with eukaryotic cells to study host–bacteria interactions. While the targeting mechanism via the Ubi29-41 peptide vector does not permit discrimination between live, dead, and metabolically inactive bacteria, the narrow emission profile of dSQ+-Ubi enables straightforward co-staining with appropriate viability markers. Furthermore, the far-red excitation and emission make dSQ+-Ubi a promising candidate for in vivo studies of bacterial infections. Moreover, its ability to label both Gram-positive and Gram-negative bacteria may be valuable for visualizing the entire host microbiota.
In addition, the clinical translation of dSQ+-Ubi should be further pursued. Although assessing the diagnostic accuracy, sensitivity, and specificity of urinary flow cytometry with dSQ+-Ubi was not the objective of the present study, it represents a logical next step in the development of dSQ+-Ubi for clinical or laboratory applications.
Author contributions
LW: conceptualization, formal analysis, investigation, software, visualization, writing – review & editing, MV: formal analysis, investigation, writing – review & editing, PW: investigation, writing – review & editing, AA: investigation, writing – review & editing, OF: investigation, writing – review & editing, NP: investigation, writing – review & editing, MF: investigation, resources, writing – review & editing, POV: methodology, resources, writing – review & editing, DD: conceptualization, formal analysis, software, writing – review & editing, DB: conceptualization, formal analysis, resources, writing – review & editing, JK: conceptualization, formal analysis, funding acquisition, investigation, methodology, supervision, visualization, writing – original draft, review and editing.Conflicts of interest
There are no conflicts to declare.Data availability
The raw data underlying this study (NMR, LC-HRMS, UV-vis absorption and fluorescence spectra, flow cytometry data, fluorescence confocal microscopy images) are openly available in Zenodo at DOI https://doi.org/10.5281/zenodo.17573344.Supplementary information (SI): Fig. S1–S3, Tables S1 and S2, synthesis and characterization of the probes, experimental details for spectroscopy, microscopy, flow cytometry, bacterial and mammalian culture. See DOI: https://doi.org/10.1039/d5sd00160a.
Acknowledgements
This work of the Strasbourg Drug Discovery and Development Institute (IMS), as part of the Interdisciplinary Thematic Institute (ITI) 2021–2028 program of the University of Strasbourg, CNRS, and Inserm, was supported by IdEx Unistra (ANR-10-IDEX-0002) and by the SFRI-STRAT'US project (ANR-20-SFRI-0012) under the framework of the French Investments for the Future Program. This work was also supported by IdEx Unistra 2022 “Recherche exploratoire”, ANR-22-CE44-0033 ULTRON, the CNRS through the MITI interdisciplinary programs “Lumière et vie” and AAP Mobility grant from the Chemical Biology Division of the French Chemical Society. L. W. was supported by a fellowship from the French Ministry of Higher Education, Research, and Innovation. We are grateful to Christel Valencia (PCBIS, UAR 3286) for providing HEK293T cells, Dr. Delphine Garnier, Dr. Estefanía Olivia and Dr. Cheng Deng (PACSI platform of PCBIS, UAR 3286) for mass spectrometry, Claudine Ebel and Muriel Philipps (IGBMC Flow Cytometry Facility, Strasbourg) for the assistance in flow cytometry experiments, Dr. Nicolas Humbert (UMR 7021 and Plateforme de Spectroscopie et de Synthèse Peptidique) for the assistance in the fluorescence spectroscopy experiments, G. Duchemin for the synthesis of LysN3-Ubi. We thank the organizers of MiFoBio 2023 (GDR ImaBio, CNRS, France) for making this collaboration possible. All experiments were performed in accordance with French bioethics laws and were approved by the Ethics Committee of the University Hospital of Saint-Étienne (reference number IRBN492025/CHUSTE). Opt-out consent was obtained from all human participants involved in this study.References
- Antimicrobial Resistance Collaborators, Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399, 629–655 CrossRef PubMed.
- F. Akram, M. Imtiaz and I. U. Haq, Emergent crisis of antibiotic resistance: A silent pandemic threat to 21st century, Microb. Pathog., 2023, 174, 105923 CrossRef CAS PubMed.
- M. A. Cook and G. D. Wright, The past, present, and future of antibiotics, Sci. Transl. Med., 2022, 14, eabo7793 CrossRef CAS PubMed.
- P. Sass, in Antibiotics, ed. P. Sass, Springer New York, New York, NY, 2017, vol. 1520, pp. 3–22 Search PubMed.
- E. D. Brown and G. D. Wright, Antibacterial drug discovery in the resistance era, Nature, 2016, 529, 336–343 CrossRef CAS PubMed.
- B. Grey, M. Upton and L. T. Joshi, Urinary tract infections: a review of the current diagnostics landscape, J. Med. Microbiol., 2023, 72, 11 CrossRef PubMed.
- M. Davenport, K. E. Mach, L. M. D. Shortliffe, N. Banaei, T.-H. Wang and J. C. Liao, New and developing diagnostic technologies for urinary tract infections, Nat. Rev. Urol., 2017, 14, 296–310 CrossRef PubMed.
- T. Bermudez, J. E. Schmitz, M. Boswell and R. Humphries, Novel technologies for the diagnosis of urinary tract infections, J. Clin. Microbiol., 2025, 63, e00306–e00324 CrossRef PubMed.
- P. Mejuto, M. Luengo and J. Díaz-Gigante, Automated Flow Cytometry: An Alternative to Urine Culture in a Routine Clinical Microbiology Laboratory?, Int. J. Microbiol., 2017, 2017, 1–8 CrossRef PubMed.
- H. M. Gilboe, O. M. Reiakvam, L. Aasen, T. Tjade, J. Bjerner, T. E. Ranheim and P. Gaustad, Rapid diagnosis and reduced workload for urinary tract infection using flowcytometry combined with direct antibiotic susceptibility testing, PLoS One, 2021, 16, e0254064 CrossRef CAS PubMed.
- B. Moshaver, F. De Boer, H. Van Egmond-Kreileman, E. Kramer, C. Stegeman and P. Groeneveld, Fast and accurate prediction of positive and negative urine cultures by flow cytometry, BMC Infect. Dis., 2016, 16, 211 CrossRef PubMed.
- E. Yusuf, B. Van Herendael and J. van Schaeren, Performance of urinalysis tests and their ability in predicting results of urine cultures: a comparison between automated test strip analyser and flow cytometry in various subpopulations and types of samples, J. Clin. Pathol., 2017, 70, 631–636 CrossRef CAS PubMed.
- O. Kocaoglu and E. E. Carlson, Progress and prospects for small-molecule probes of bacterial imaging, Nat. Chem. Biol., 2016, 12, 472–478 CrossRef CAS PubMed.
- B. Mills, M. Bradley and K. Dhaliwal, Optical imaging of bacterial infections, Clin. Transl. Imaging, 2016, 4, 163–174 CrossRef PubMed.
- A. A. Ordonez, M. A. Sellmyer, G. Gowrishankar, C. A. Ruiz-Bedoya, E. W. Tucker, C. J. Palestro, D. A. Hammoud and S. K. Jain, Molecular imaging of bacterial infections: Overcoming the barriers to clinical translation, Sci. Transl. Med., 2019, 11, eaax8251 CrossRef PubMed.
- Y. Huang, W. Chen, J. Chung, J. Yin and J. Yoon, Recent progress in fluorescent probes for bacteria, Chem. Soc. Rev., 2021, 50, 7725–7744 RSC.
- J. Zhan, Y. Cai, P. Cheng, L. Zheng and K. Pu, Body fluid diagnostics using activatable optical probes, Chem. Soc. Rev., 2025, 54, 3906–3929 RSC.
- M. Íñigo, A. Coello, G. Fernández-Rivas, M. Carrasco, C. Marcó, A. Fernández, T. Casamajor and V. Ausina, Evaluation of the SediMax automated microscopy sediment analyzer and the Sysmex UF-1000i flow cytometer as screening tools to rule out negative urinary tract infections, Clin. Chim. Acta, 2016, 456, 31–35 CrossRef PubMed.
- T. Parasassi, G. De Stasio, A. d'Ubaldo and E. Gratton, Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence, Biophys. J., 1990, 57, 1179–1186 CrossRef CAS PubMed.
- L. Weiss, A. Mirloup, L. Blondé, H. Manko, J. Peluso, D. Bonnet, D. Dziuba and J. Karpenko, Fluorescent Antimicrobial Peptides Based on Nile Red: Effect of Conjugation Site and Chemistry on Wash-free Staining of Bacteria, Bioconjugate Chem., 2024, 35(11), 1779–1787 CrossRef CAS PubMed.
- A. R. Akram, N. Avlonitis, A. Lilienkampf, A. M. Perez-Lopez, N. McDonald, S. V. Chankeshwara, E. Scholefield, C. Haslett, M. Bradley and K. Dhaliwal, A labelled-ubiquicidin antimicrobial peptide for immediate in situ optical detection of live bacteria in human alveolar lung tissue, Chem. Sci., 2015, 6, 6971–6979 RSC.
- A. Baibek, M. Üçüncü, E. A. Blackburn, M. Bradley and A. Lilienkampf, Wash-free, peptide-based fluorogenic probes for microbial imaging, Pept. Sci., 2021, 113(1), e24167–e24174 CrossRef CAS.
- J. T. Mika, A. J. Thompson, M. R. Dent, N. J. Brooks, J. Michiels, J. Hofkens and M. K. Kuimova, Measuring the Viscosity of the Escherichia coli Plasma Membrane Using Molecular Rotors, Biophys. J., 2016, 111, 1528–1540 CrossRef CAS PubMed.
- P. Loison, N. A. Hosny, P. Gervais, D. Champion, M. K. Kuimova and J.-M. Perrier-Cornet, Direct investigation of viscosity of an atypical inner membrane of Bacillus spores: a molecular rotor/FLIM study, Biochim. Biophys. Acta, 2013, 1828, 2436–2443 CrossRef CAS PubMed.
- L. Ding, X. Wang, J. Wang, H. Wang, L. Yu, J. Liu, J. Yu, T. Xue, X. Yang and L. Xue, Fluorogenic Probes for Real-Time Tracking of Bacterial Cell Wall Dynamics with Nanoscopy, ACS Nano, 2025, 19, 14389–14403 CrossRef CAS PubMed.
- C. Wang, J. Wang, K. Xue, M. Xiao, K. Wu, S. Lv, B. Hao and C. Zhu, Polarity-Sensitive Fluorescent Probe for Reflecting the Packing Degree of Bacterial Membrane Lipids, Anal. Chem., 2022, 94, 3303–3312 CrossRef CAS PubMed.
- A. P. Marshall, J. D. Shirley and E. E. Carlson, Enzyme-targeted fluorescent small-molecule probes for bacterial imaging, Curr. Opin. Chem. Biol., 2020, 57, 155–165 CrossRef CAS PubMed.
- A. S. Klymchenko, Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications, Acc. Chem. Res., 2017, 50, 366–375 CrossRef CAS PubMed.
- F. Bouhedda, K. T. Fam, M. Collot, A. Autour, S. Marzi, A. Klymchenko and M. Ryckelynck, A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells, Nat. Chem. Biol., 2020, 16, 69–76 CrossRef CAS PubMed.
- A. Okamoto, ECHO probes: a concept of fluorescence control for practical nucleic acid sensing, Chem. Soc. Rev., 2011, 40, 5815 RSC.
- I. A. Karpenko, M. Collot, L. Richert, C. Valencia, P. Villa, Y. Mély, M. Hibert, D. Bonnet and A. S. Klymchenko, Fluorogenic Squaraine Dimers with Polarity-Sensitive Folding As Bright Far-Red Probes for Background-Free Bioimaging, J. Am. Chem. Soc., 2015, 137, 405–412 CrossRef CAS PubMed.
- L. Esteoulle, F. Daubeuf, M. Collot, S. Riché, T. Durroux, D. Brasse, P. Marchand, I. A. Karpenko, A. S. Klymchenko and D. Bonnet, A near-infrared fluorogenic dimer enables background-free imaging of endogenous GPCRs in living mice, Chem. Sci., 2020, 11, 6824–6829 RSC.
- Y. Berthomé, J. Gerber, F. Hanser, S. Riché, N. Humbert, C. Valencia, P. Villa, J. Karpenko, O. Florès and D. Bonnet, Rational Design of Cyanine-Based Fluorogenic Dimers to Reduce Nonspecific Interactions with Albumin and Lipid Bilayers: Application to Highly Sensitive Imaging of GPCRs in Living Cells, Bioconjugate Chem., 2024, 35, 1182–1189 CrossRef PubMed.
- K. T. Fam, M. Collot and A. S. Klymchenko, Probing biotin receptors in cancer cells with rationally designed fluorogenic squaraine dimers, Chem. Sci., 2020, 11, 8240–8248 RSC.
- K. T. Fam, L. Saladin, A. S. Klymchenko and M. Collot, Confronting molecular rotors and self-quenched dimers as fluorogenic BODIPY systems to probe biotin receptors in cancer cells, Chem. Commun., 2021, 57, 4807–4810 RSC.
- I. O. Aparin, R. Yan, R. Pelletier, A. A. Choi, D. I. Danylchuk, K. Xu and A. S. Klymchenko, Fluorogenic Dimers as Bright Switchable Probes for Enhanced Super-Resolution Imaging of Cell Membranes, J. Am. Chem. Soc., 2022, 144, 18043–18053 CrossRef CAS PubMed.
- C. Moore, R. M. Borum, Y. Mantri, M. Xu, P. Fajtová, A. J. O'Donoghue and J. V. Jokerst, Activatable Carbocyanine Dimers for Photoacoustic and Fluorescent Detection of Protease Activity, ACS Sens., 2021, 6, 2356–2365 CrossRef CAS PubMed.
- D. Yao, Z. Lin and J. Wu, Near-Infrared Fluorogenic Probes with Polarity-Sensitive Emission for in Vivo Imaging of an Ovarian Cancer Biomarker, ACS Appl. Mater. Interfaces, 2016, 8, 5847–5856 CrossRef CAS PubMed.
- J. Bhatt Mitra, V. K. Sharma, A. Mukherjee, V. Garcia Sakai, A. Dash and M. Kumar, Ubiquicidin-Derived Peptides Selectively Interact with the Anionic Phospholipid Membrane, Langmuir, 2020, 36, 397–408 CrossRef CAS PubMed.
- C. P. J. M. Brouwer, S. J. P. Bogaards, M. Wulferink, M. P. Velders and M. M. Welling, Synthetic peptides derived from human antimicrobial peptide ubiquicidin accumulate at sites of infections and eradicate (multi-drug resistant) Staphylococcus aureus in mice, Peptides, 2006, 27, 2585–2591 CrossRef CAS PubMed.
- A. R. Akram, N. Avlonitis, E. Scholefield, M. Vendrell, N. McDonald, T. Aslam, T. H. Craven, C. Gray, D. S. Collie, A. J. Fisher, P. A. Corris, T. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Enhanced avidity from a multivalent fluorescent antimicrobial peptide enables pathogen detection in a human lung model, Sci. Rep., 2019, 9, 8422 CrossRef PubMed.
- M. M. Welling, A. Bunschoten, J. Kuil, R. G. H. H. Nelissen, F. J. Beekman, T. Buckle and F. W. B. van Leeuwen, Development of a Hybrid Tracer for SPECT and Optical Imaging of Bacterial Infections, Bioconjugate Chem., 2015, 26, 839–849 CrossRef CAS PubMed.
- C. Liu and Y. Gu, Noninvasive optical imaging of Staphylococcus aureus infection in vivo using an antimicrobial peptide fragment based near-infrared fluorescent probes, J. Innovative Opt. Health Sci., 2013, 06, 1350026 CrossRef.
- H. Chen, C. Liu, D. Chen, K. Madrid, S. Peng, X. Dong, M. Zhang and Y. Gu, Bacteria-Targeting Conjugates Based on Antimicrobial Peptide for Bacteria Diagnosis and Therapy, Mol. Pharmaceutics, 2015, 12, 2505–2516 CrossRef CAS PubMed.
- K. Ilina, W. M. MacCuaig, M. Laramie, J. N. Jeouty, L. R. McNally and M. Henary, Squaraine Dyes: Molecular Design for Different Applications and Remaining Challenges, Bioconjugate Chem., 2020, 31, 194–213 CrossRef CAS PubMed.
- J. Karpenko, A. S. Klymchenko, S. Gioria, R. Kreder, I. Shulov, P. Villa, Y. Mély, M. Hibert and D. Bonnet, Squaraine as a bright, stable and environment-sensitive far-red label for receptor-specific cellular imaging, Chem. Commun., 2015, 51, 2960–2963 RSC.
- M. L. Wilson and L. Gaido, Laboratory Diagnosis of Urinary Tract Infections in Adult Patients, Clin. Infect. Dis., 2004, 38, 1150–1158 CrossRef PubMed.
- Y. Zhang, X. Yue, B. Kim, S. Yao and K. D. Belfield, Deoxyribonucleoside-Modified Squaraines as Near-IR Viscosity Sensors, Chem. – Eur. J., 2014, 20, 7249–7253 CrossRef CAS PubMed.
- S. Pfister, J. Lesieur, P. Bourdoncle, M. Elhassan, P. Didier, N. Anton, H. Anton and M. Collot, Red-Emitting Pyrrolyl Squaraine Molecular Rotor Reports Variations of Plasma Membrane and Vesicular Viscosity in Fluorescence Lifetime Imaging, Anal. Chem., 2024, 96, 12784–12793 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |