 Open Access Article
Open Access ArticleRecent advances in phenotypic antimicrobial susceptibility testing enabled by microfluidic technologies
Mo
Shen
ab,
Qi
Wang
d,
Qingqing
Luo
b,
Jiatong
Zhao
e and
Feng
Shen
 *abc
*abc
aSixth People's Hospital, School of Medicine & School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200030, PR China
bSchool of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200030, China. E-mail: feng.shen@sjtu.edu.cn
cZhengzhou Industrial Technology Research Institute of Shanghai Jiao Tong University, Zhengzhou 450016, China
dRuijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200025, China
eDepartment of biology Brandeis University, Waltham, MA 02453, USA
First published on 3rd November 2025
Abstract
Antimicrobial resistance (AMR) poses an urgent global health threat, driving the need for rapid and accurate antimicrobial susceptibility testing (AST). Traditional phenotypic AST methods remain the clinical gold standard but are hindered by prolonged turnaround times and labor-intensive procedures. Microfluidic technologies have emerged as transformative platforms, enabling miniaturized, high-throughput, and integrated phenotypic AST workflows with accelerated result delivery. This review comprehensively summarizes recent advances in microfluidic phenotypic AST, categorizing platforms by cultivation strategies—such as static chambers, flow chambers, SlipChip variants, and hybrid droplet-chamber systems—and surveying diverse signal detection modalities including fluorescence, label-free imaging, Raman, electrical, and mechanical readouts, each offering distinct advantages and limitations. Key innovations such as concentration gradient generation, digital single-cell manipulation, and AI-enhanced image analysis have significantly improved sensitivity, speed, and clinical applicability. However, widespread adoption remains challenged by sample-to-result integration, slow-growing pathogens, interference from residual antibiotics, and the lack of robust standardization. We further discuss emerging solutions, including automated sample preparation, multimodal detection, and computational data fusion, and outline future opportunities for translating microfluidic phenotypic AST into routine diagnostics. Collectively, these advances hold substantial promise for combating AMR by enabling personalized, rapid, and actionable antimicrobial therapy.
1. Introduction
Antimicrobial resistance (AMR) is an escalating global health crisis, currently associated with over 1.27 million deaths each year and projected to result in tens of millions of fatalities annually by 2050 if not effectively addressed.1,2 In routine clinical microbiology, pathogens are isolated from the patient specimen and prepared as a normalized inoculum (e.g., against a turbidity standard). The inoculum is then exposed to a twofold serial dilution panel of antibiotics in broth (broth microdilution) or on solid medium (agar dilution), or challenged on a lawn with paper disks. After incubation, visible growth is assessed to determine the minimum inhibitory concentration (MIC) or inhibition zone diameters, which are then translated into susceptible/intermediate/resistant (S/I/R) categories according to CLSI/EUCAST criteria. While these approaches remain clinical gold standards due to their established protocols and broad regulatory acceptance, they are inherently slow, typically requiring 24 to 48 hours to generate results, and sometimes even longer for certain slow-growing pathogens. This prolonged turnaround time significantly delays clinical decision-making and often leads to empirical—and sometimes inappropriate—antibiotic use, further accelerating the spread of resistance.3–5 These delays have especially detrimental effects in severe infections, where timely antimicrobial intervention is crucial for optimal patient outcomes.6,7To overcome workflow bottlenecks and improve efficiency, several automated AST systems such as VITEK® 2 and BD Phoenix™ have been developed. These platforms automate sample processing and result interpretation, offering simplified operation and increased throughput compared with conventional manual methods. However, despite these advances, most automated systems still require 6–24 hours to generate results and remain limited in their capacity for rapid, comprehensive susceptibility profiling. Consequently, there is an urgent need for next-generation AST technologies capable of providing faster and more actionable results to support timely clinical decision-making and combat the ongoing AMR crisis.
Although molecular methods, which detect specific molecular biomarkers of resistance are emerging, they may not comprehensively predict actual clinical resistance phenotypes due to the complexity of resistance mechanisms and their variable expression in clinical isolates.8,9 Phenotypic AST methods thus offer a more accurate representation of how pathogens will respond to specific antibiotics in clinical settings, better informing clinical decisions and enabling personalized antimicrobial therapy.10–12
Microfluidic technologies have recently emerged as promising approaches for rapid AST, offering distinct advantages over conventional methods, such as significantly reduced reaction volumes, accelerated mass and heat transfer, precise fluid control, high-throughput capabilities, single-cell resolution, and potential integration with automated and portable diagnostic systems.13–15 Microfluidics handles picoliter–microliter volumes in microscale channels, where laminar flow and high surface-to-volume ratios make transport fast and predictable, enabling precise dosing, compartmentalization, and tight control of antibiotic exposure—ideal conditions for rapid phenotypic AST.
Microfluidic phenotypic AST platforms can be categorized based on cultivation methods into static chambers, continuous flow chambers, droplet-based systems, and SlipChip technologies, each exhibiting unique strengths in throughput, assay flexibility, and integration potential.16–19 Recent advances in signal detection techniques, such as optical microscopy, fluorescence imaging, Raman spectroscopy, electrical impedance measurements, and electrochemical sensors, have considerably enhanced the sensitivity and rapidity of phenotypic AST. These technologies now enable determination of antibiotic susceptibility within hours, or even minutes, compared to the days required by traditional methods, markedly improving their clinical applicability.11,20–23 Nevertheless, clinical implementation of microfluidic AST still encounters several critical challenges, including complex sample matrices, extensive sample preparation requirements, interference from residual antibiotics in clinical specimens, and the necessity for robust standardization and regulatory approvals.24,25
In this review, we provide a design-oriented analysis of phenotypic microfluidic AST, including device design, cultivation architectures, and detection modalities. Recent reviews have surveyed microfluidic AST from different angles: a 2022 Lab on a Chip critical review catalogued phenotypic and pheno-molecular approaches across static chambers, flow systems, droplet platforms, and pheno-molecular assays, emphasizing bottlenecks such as multiplexing and detection time and outlining commercialization considerations;13 an Accounts of Chemical Research review focused on droplet microfluidics, single-cell/digital analyses, and practical challenges such as dye leakage and end-point vs. kinetic readouts;11 and a recent Biosensors & Bioelectronics review highlighted colorimetric microfluidic AST platforms aimed at point-of-care use.26 Distinct from these contributions, this review explicitly links cultivation architectures (static, flow, SlipChip, hybrid droplet-chamber) with detection modalities (fluorescence, label-free optical, Raman/SERS, electrical, mechanical), extracts comparable operational metrics (antibiotic exposure time, end-to-end turnaround, sample compatibility), and embeds practical “how-to” guidance for sample-to-result integration, slow-grower analysis, and mitigation of residual-antibiotic interference. We also outline opportunities with AI and integrated workflows to support clinical translation and reduce inappropriate antibiotic use.27,28
2. Cultivation methods in phenotype microfluidic AST
The selection and optimization of cultivation methods are fundamental to the effectiveness and applicability of phenotypic microfluidic AST. This section provides a detailed overview of various cultivation strategies currently employed in microfluidic AST systems, including static chambers, continuous flow chambers, hybrid droplet-chamber devices, and SlipChip platforms. Each method offers distinct operational characteristics, performance advantages, and practical considerations, influencing the choice of technique based on the specific clinical or laboratory requirements. We discuss the principles underlying these cultivation methods, their practical implementation, as well as their respective strengths, providing insights into how they contribute to achieving rapid, accurate, and clinically relevant AST outcomes. For side-by-side comparison, Table 1 compares four representative cultivation architectures used in phenotypic microfluidic AST—microfluidic static chambers, SlipChip systems, microfluidic flow chambers, and hybrid droplet-chamber systems. For each class, the table summarizes the core principle, key operational characteristics, practical advantages, and typical application scenarios.| Cultivation method type | Core principle | Operational characteristics | Advantages | Ref. |
|---|---|---|---|---|
| Microfluidic static chambers | No active fluid flow; antibiotic-bacteria contact via diffusion; chamber arrays enable parallel culture | No pumps or valves needed; low sample volume concentration gradients via centrifugation or diffusion | Low operation threshold for clinical labs; controllable cost for mass production; high environmental stability | 34–37, 16 |
| SlipChip systems | Two chips with complementary microstructures slide to seal chambers, mix reagents, and generate gradients; no external drive | One-step sample distribution; low cross-contamination risk; integrable with culture and detection functions | High reagent utilization (nL volume); supports multi-antibiotic detection; low leakage risk | 43, 44, 17 |
| Microfluidic flow chambers | Continuous fluid perfusion for nutrient supply and waste removal; simulates in vivo environment; stable gradients | Pump-controlled flow; real-time antibiotic adjustment; supports long-term biofilm culture | In vivo-like conditions; direct use of blood samples | 14, 47–51 |
| Hybrid droplet-chamber systems | Oil-isolated droplets encapsulate single cells or antibiotics; chambers fix droplets for detection | Automated workflow; digital single-cell analysis; low reagent volume | Ultra-high throughput for large-scale screening; captures heterogeneous resistance; low contamination | 11, 27, 56 |
2.1 Microfluidic static chambers for quantitative AST
Microfluidic static chambers are defined by the absence of active fluid flow during bacterial cultivation, enabling controlled and stable microenvironments for observing antibiotic responses.29,30 Typically, these devices consist of multiple isolated reaction chambers or wells for defined reagent loading and optical monitoring of growth.31 The static layout reduces fluidic complexity and supports parallel assays, which can be aligned with routine laboratory workflows.32,33Ma et al. utilized a polymer-based lab-on-a-chip with arrayed reaction chambers. Each chamber was preloaded with colorimetric media and antibiotics, allowing visual or optical readout of Campylobacter spp. growth and susceptibility in a short timeframe.34 The simple chamber layout supports direct visual interpretation and straightforward operation for field and clinical use. Wang and Erickson developed a paper-based static-chamber device using wax-printed microchannels to form defined reaction zones. Antibiotics and resazurin were dried in situ, so a single capillary driven loading step initiated the test. The format is portable and cost-conscious, enabling visual growth calls and semi-quantitative MIC in a short timeframe, which is practical for resource-limited settings.35
In a related approach, Azizi et al. introduced a gradient-based static microfluidic platform composed of parallel channels separated by oil barriers. Diffusion driven concentration gradients across adjacent chambers enabled controlled antibiotic dosing, and the clear chamber geometry facilitated fluorescence imaging for MIC determination in a short timeframe.36 Nguyen et al. leveraged a ladder-shaped centrifugal microfluidic design that generates a standardized twofold serial dilution gradient (Fig. 1A). The structure automates fluid handling and yields reproducible AST readouts in a short timeframe.16 The use of centrifugal forces simplifies sample movement and metering. Li et al. further advanced the static-chamber concept with an under-oil open microfluidic system. Chambers formed by patterned hydrophilic regions and covered by an oil layer mitigate evaporation and contamination while maintaining direct optical access for single-cell imaging (Fig. 1B). MIC assessment can be completed in a short timeframe, which streamlines workflow and offers operational flexibility.22
 | ||
| Fig. 1 AST using static chambers. (A) Illustration of a ladder microfluidic AST system, featuring a structure with microchamber triplicates, a serpentine mixer, and side channel hydraulic resistors; the workflow includes sequential loading of bacterial suspension and antibiotic/culture medium to generate an on-chip exponential concentration gradient, oil-based isolation of microchambers, and resazurin fluorescence readout for MIC determination. Reproduced with permission from ref. 16. Copyright 2023, Springer Nature. (B) Configuration and working principle of the UOMS-AST platform, featuring a chambered coverglass patterned for an array of sessile microdrops under oil with surface energy confinement, where bacterial suspensions (with or without antimicrobial) are inoculated, and antimicrobial susceptibility is assessed by live-cell imaging of bacterial confluency.22 Reproduced with permission from ref. 22. Copyright 2023, Royal Society of Chemistry. (C) Schematic diagram of CCM for AST. The entire on-chip process, which includes sample loading, centrifugation, and data acquisition, typically takes 4–9 h to generate AST results. Reproduced with permission from ref. 37. Copyright 2024, Elsevier. | ||
Building on similar centrifugal principles, Pang et al. designed a controlled-diffusion centrifugal platform with radial reaction chambers on a disk. Antibiotic gradients are generated by controlled diffusion under rotation, and samples are transferred into isolated chambers automatically (Fig. 1C). In combination with a mobile detection module, the device supports MIC calls on a several hour timescale, improving portability and user friendliness.37 The integration of automated handling with compact detection indicates potential for near-field deployment, pending further evaluation against standardized clinical endpoints.
2.2 SlipChip for quantitative AST
SlipChip technology manipulates fluids by the relative sliding of two plates patterned with complementary microstructures.38,39 Although it was not originally developed for AST,40 SlipChip platforms have been adapted to this application by leveraging static-chamber principles.41 In AST workflows, SlipChip effectively functions as a static array that isolates microvolumes without active flow, while sliding enables rapid partitioning, controlled mixing, and convenient formation of antibiotic concentration series. This architecture supports parallel testing and shows potential for portable or near-patient use at the level of principle.Cai et al., utilized a two-step sliding mechanism to integrate cell culture, lysis, and enzymatic signal generation in isolated chambers. This platform enabled label-free Escherichia coli (E. coli) detection with a detection limit as low as 8 CFU per chamber within 5 hours, using a simple polydimethylsiloxane (PDMS)-based design compatible with standard microplate readers. The chip's ability to compartmentalize reactions without pumps or valves highlighted its potential for low-cost, decentralized diagnostics.42 Li et al. introduced a combinatorial screening SlipChip that enabled high-throughput phenotypic AST by guiding bacterial suspensions into preloaded chambers containing dried antibiotics (Fig. 2A). Utilizing a bead-like “pearl-chain” channel structure, the chip partitions 50 nL droplets into 192 microchambers with minimal cross-contamination. MIC calls can be obtained in a short timeframe by bright-field imaging, with trends consistent with commercial systems, while improving reagent efficiency and scalability for laboratory workflows.43
 | ||
| Fig. 2 AST using SlipChip. (A) Principle and workflow of the cs-SlipChip, featuring a two-plate structure with preloaded antibiotics in microwells; bacterial solution is loaded, manually partitioned into droplets that mix with different antibiotics, enabling rapid on-chip antimicrobial susceptibility profiling. Reproduced with permission from ref. 42. Copyright 2022, Royal Society of Chemistry. (B) Mechanism and workflow of the gradient droplet SlipChip (gd-SlipChip), featuring a two-plate design that enables formation of antibiotic concentration gradients and gradient droplets via sequential loading, diffusion, and manual slipping steps for on-chip multiplexed reagent delivery and analysis. Reproduced with permission from ref. 44. Copyright 2022, American Chemical Society. (C) Principle and workflow of the nd-SlipChip, showing a dual-plate design with microwells and loading channels for automated serial dilution, uniform droplet generation. Reproduced with permission from ref. 17. Copyright 2025, Elsevier. | ||
Further advancing the approach, Liu et al. developed a self-partitioning gradient-droplet SlipChip that relies on molecular diffusion and geometry-guided splitting to automatically produce a wide-range concentration gradient within a droplet array (Fig. 2B). This enables gradient-based AST across multiple concentrations in parallel, without external pumps or complex control. Using clinical Escherichia coli strains against nitrofurantoin, results were obtained on a several-hour timescale and were in good agreement with standard methods.44
More recently, Wang et al. reported a nano-dilution SlipChip combining sequential sliding with wet-etching to generate small-volume gradient arrays (Fig. 2C). The platform supports AST, combination testing, and phage assays on a single chip, providing MIC determinations in a short timeframe. Its performance was validated against 24 clinical strains with 97.9% concordance with broth microdilution (BMD) gold standards. Furthermore, the nd-SlipChip successfully analyzed clinical urine samples with minimal preprocessing (e.g., filtration and centrifugation), highlighting its real-world clinical applicability and promise for personalized medicine.17
The aforementioned studies illustrate the trajectory of SlipChip in AST from early feasibility to integrated, high-throughput formats. By integrating the spatial control of static chambers with simple, on-chip manipulation, SlipChip offers a practical route to parallel phenotypic testing while keeping operation straightforward.
2.3 Microfluidic flow chambers for quantitative AST
Microfluidic flow chambers provide a continuous fluid environment for bacterial cultivation and antibiotic exposure, distinguishing them from static systems by enabling dynamic control over reagent supply, waste removal, and real-time monitoring.7,9,45 These systems typically involve the perfusion of media or antibiotic-containing solutions through microchannels or flow-through reaction chambers, creating a physiologically relevant and tunable microenvironment. Flow chambers are particularly advantageous for capturing rapid bacterial responses, establishing stable concentration gradients, and integrating with real-time imaging or biosensing modalities.7,45 Unlike static systems, flow-based platforms more closely mimic in vivo conditions by maintaining a constant nutrient supply and removing inhibitory metabolites, thereby supporting more precise and accelerated phenotypic AST.46,47 Consequently, microfluidic flow chambers have emerged as powerful tools for conducting high-resolution, time-resolved studies of bacterial growth dynamics, enabling faster and more sensitive antimicrobial susceptibility evaluations.Recent developments illustrate diverse flow-based architectures. Zhu et al. designed an integrated chip that couples a density-based bacterial separation module with an array of flow-through AST chambers14 (Fig. 3A). Bacteria are first enriched from whole blood (centrifugal separation) and then distributed into parallel antibiotic-loaded channels via vacuum-assisted self-filling. Continuous perfusion supports rapid response measurement and enables direct-from-blood AST within a short timeframe. Wistrand-Yuen et al. employed a dual laminar flow design flanking gel-confined growth chambers, where lateral diffusion of antibiotics forms stable concentration gradients.48 The platform preserves distinct flow boundaries while sustaining oxygen and nutrient diffusion through the gel matrix, allowing high-throughput, image-based MIC determination on a several hour timescale. Blanco-Cabra et al. introduced the BiofilmChip, addressing biofilm-associated resistance under flow conditions49 (Fig. 3B). Interdigitated electrodes embedded in the flow chambers, together with a stable perfusion loop, enable real-time impedance monitoring of biofilm biomass changes to infer susceptibility. A pre-chamber stabilizes flow and mitigates shear during manual inoculation, helping to achieve homogeneous biofilm formation across chambers; this expands AST evaluation beyond planktonic cells.
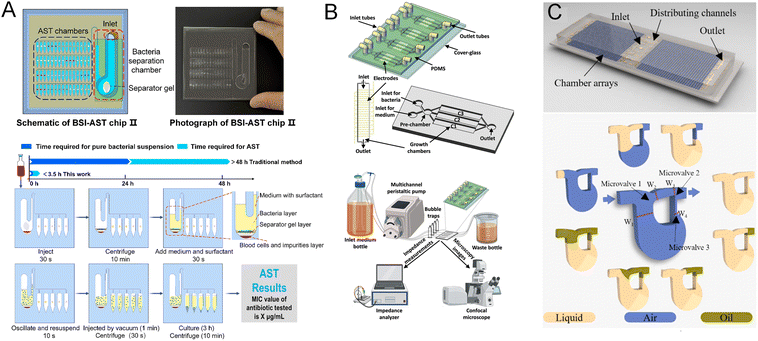 | ||
| Fig. 3 AST using flow chambers. (A) Schematic and workflow of the BSI-AST chip enabling rapid AST directly from positive blood cultures, featuring direct processing without subculture and completing bacterial separation and susceptibility testing in under 3.5 hours. Reproduced with permission from ref. 14. Copyright 2023, American Chemical Society. (B) BiofilmChip platform for real-time biofilm monitoring, showing the 3D chip design with integrated interdigitated electrodes, single and multi-chamber views, and the complete experimental setup enabling controlled medium perfusion and electrochemical monitoring of biofilm growth and treatment. Reproduced with permission from ref. 49. Copyright 2021, Springer Nature. (C) Self-priming digital microfluidic AST chip, featuring an array of 1824 chambers with integrated microvalves for pump-free sample loading, enabling customizable on-chip antibiotic pre-coating and rapid, single-cell level MIC determination. Reproduced with permission from ref. 52. Copyright 2025, Elsevier. | ||
Electrochemical integration in flow channels was further refined by Jeon et al., who used a branched layout that automatically mixes bacteria with antibiotics to generate five-step serial dilutions.50 Continuous flow partitions samples across test chambers and supports dynamic growth measurement by tracking double-layer capacitance on gold electrodes, providing MIC calls on the order of a few hours. Pang et al. engineered a multiplexed, vacuum-driven flow chip with eight main channels feeding forty nanoliter-scale chambers preloaded with antibiotics.51 Upon sample introduction, oil-phase segmentation creates sealed compartments to improve isolation and reduce evaporation/cross-talk (Fig. 3C). This balances flow-based reagent control with discrete-chamber operation and yields MIC assessments in a short timeframe consistent with conventional dilution readouts in their tests. A subsequent iteration by the same group extended the concept to single-cell resolution using digital microfluidics.52 More than 1800 reaction units receive metered volumes by self-priming capillary action, and oil segmentation preserves distribution while enabling stochastic single-cell loading; resazurin-based readouts support MIC calls on a several-hour timescale.
Collectively, these studies demonstrate that microfluidic flow chambers consistently offer: (1) continuous medium renewal and by-product removal to support viability and responsive growth; (2) the ability to form and maintain stable chemical gradients via lateral diffusion or serial/segmented flow; and (3) compatibility with real-time biosensing and imaging.
2.4 Hybrid droplet-chamber devices for quantitative AST
Hybrid droplet-chamber devices represent a novel class of microfluidic AST platforms that combine the compartmentalization advantages of droplet microfluidics with the spatial organization and controllability of chamber-based designs.53,54 These systems typically integrate droplet generation modules with structured microchambers or arrays, enabling the encapsulation of bacterial samples into discrete, physically isolated reaction units.55 The use of oil phase segmentation enables stable, leak-free isolation of test conditions, effectively preventing cross-contamination and supporting long-term cultivation or observation. Meanwhile, the physical layout of chambers facilitates straightforward imaging, reagent loading, and downstream analysis, overcoming limitations of pure droplet in flow systems, which are more challenging to track and manipulate individually. By leveraging droplet-based metering, hybrid devices achieve high-throughput screening of multiple antibiotics or concentrations in parallel, while the chamber structure provides enhanced environmental control, optical access, and standardization. These hybrid platforms are particularly well-suited for applications requiring single-cell resolution, concentration gradient formation, or combinatorial testing, making them a promising solution for future scalable, rapid, and accurate phenotypic AST.In recent developments, Graf et al. further advanced this concept by integrating deep learning into the analytical pipeline.77 They generated picoliter droplets through flow focusing to encapsulate single bacterial cells, which were subsequently analyzed using angle-resolved light scattering. A convolutional neural network (CNN) classified the growth status of each droplet, enabling label-free, rapid MIC determination within a few hours. This combination of microfluidic droplet control with intelligent image analysis exemplifies the potential of computational tools to enhance microfluidic AST performance.
Building on gradient integration strategies, Kim et al. designed a sophisticated PDMS-based hybrid chip that automates every step from antibiotic gradient formation to droplet generation and incubation. Eight concentration levels are generated in parallel and encapsulated into thousands of droplets, each seeded with individual bacteria,27 specific method shown in Fig. 4A. These droplets are incubated and imaged using phase contrast microscopy, with automated image processing providing precise growth quantification. The device achieves label-free, multiplexed AST at single-cell resolution, showcasing the power of microfluidics for automating complex cultivation protocols. Azizi et al. proposed an approach by using egg-shaped multivolume microchambers that inherently generate antibiotic concentration gradients via differential diffusion,56 eliminating the need for external gradient generators (Fig. 4B). Each chamber is isolated by an oil phase after loading, preventing cross-contamination. This design uniquely exploits variable chamber volumes to induce diffusion-driven gradients in a compact, pump-free layout. The result is a highly accessible and low-complexity platform that still supports accurate and rapid MIC determination across multiple antibiotics.
 | ||
| Fig. 4 AST using hybrid droplet-chamber. (A) Schematic of microfluidic AST workflow, illustrating a network of eight flow-focusing generators for antibiotic concentration gradient formation, encapsulation and incubation of bacteria in droplets, and subsequent MIC determination based on droplet generation and image analysis. Reproduced with permission from ref. 27. Copyright 2024, Royal Society of Chemistry. (B) Principles and workflow of the EL-MVM2 platform, featuring an egg-inspired microchamber design for in situ generation of antibiotic concentration gradients, sequential sample and drug loading, oil-washing, and homogeneous drug distribution for microfluidic AST. Reproduced with permission from ref. 56. Copyright 2021, American Chemical Society. | ||
When considered as a whole, these studies demonstrate how hybrid droplet-chamber systems combine the strengths of droplet compartmentalization with the spatial control of chambers, enabling high-throughput, low-volume, and highly parallel AST. Innovations such as automated gradient integration, digital single-cell encapsulation, diffusion-driven passive gradients, and AI-enhanced image analysis are advancing this format toward practical clinical deployment.
3. Signal detection in phenotypic microfluidic AST
Signal detection methods are fundamental to the performance and practicality of phenotypic microfluidic AST platforms. While the cultivation strategy determines how bacteria interact with antibiotics, it is the detection modality that ultimately defines how quickly, accurately, and sensitively bacterial responses can be interpreted.57,58 The miniaturization of microfluidic systems presents unique challenges for signal readout, including reduced optical path lengths, limited sample volumes, and the need for high spatial and temporal resolution.59,60 Consequently, a wide variety of signal detection strategies have been adopted to address these challenges. Based on the classification in this review, these methods can be grouped into three primary categories: optical detection, electrical detection, and mechanical detection (Table 2). These categories represent the mainstream techniques used to monitor bacterial growth and activity in microfluidic AST platforms. Each strategy exhibits distinct strengths and limitations in terms of sensitivity, throughput, instrumentation requirements, and suitability for point-of-care testing.| Types of methods | Detection methods | Core detection mechanism | Technical feature | Ref. |
|---|---|---|---|---|
| Optical detection | Fluorescent markers | Specific interaction between fluorescent dyes and bacterial components (cell membrane, metabolites, nucleic acids) to reflect viability or quantity | High sensitivity; multiplexing; single-cell resolution; compatible with many dyes and reporters | 64, 68, 69, 72, 73 |
| Optical imaging without labeling | Quantifies bacterial growth via optical signal changes (morphology, light scattering, autofluorescence) without labels | Label-free; preserves native physiology; fast and automatable; suitable for high-throughput analysis | 76–78 | |
| Raman-based AST | Uses molecular vibration fingerprints to distinguish susceptibility; SERS enhances signals for single-cell detection | Label-free and chemically specific; single-cell/single-droplet analysis; rapid detection | 81, 82 | |
| Electrical detection | Electrical impedance | Detects changes in medium electrical properties (resistance, dielectric constant) caused by bacterial growth | Label-free; real-time; rapid; high-throughput and automatable; suitable for static chambers and single-cell analysis | 87–90 |
| Electrochemical | Reflects bacterial activity via electrochemical reactions (redox, capacitance change, electricity production) between metabolism and electrodes | Simple, low-cost hardware; label-free; suitable for multiplexing; compatible with disposable chips | 50, 91–93 | |
| Mechanical detection | Cantilever-based sensors | Judges susceptibility via cantilever vibration/displacement changes induced by bacterial activity | Ultra-sensitive; growth-independent; fast; label-free and suitable for slow-growing or dormant pathogens | 23, 94 |
Importantly, detection methods must be compatible with the physical architecture of the microfluidic device and the time constraints of clinical decision-making. High-resolution optical imaging offers excellent spatial information and is particularly useful in single-cell AST systems, but may require sophisticated instrumentation. In contrast, electrical and colorimetric approaches offer simpler integration and faster signal acquisition, making them well-suited for point-of-care applications. Increasingly, real-time and automated detection systems are being developed to improve throughput and objectivity. Advanced computational tools, including machine learning and image processing algorithms, are now frequently employed to analyze signal patterns, particularly in high-content or label-free systems. This chapter categorizes and evaluates major signal detection strategies in microfluidic AST platforms, with emphasis on their technical principles, implementation compatibility, sensitivity, speed, and potential for clinical translation.
3.1 Optical detection for phenotypic microfluidic AST
Nguyen et al. developed a microfluidic chip that uses resazurin, a metabolic dye, to rapidly determine bacterial susceptibility.33 Upon reduction by viable bacterial cells, resazurin is converted into fluorescent resorufin, enabling quantification of bacterial viability under different antibiotic conditions. Their system integrates a diffusive convective loading mechanism to generate an antibiotic concentration gradient within nanoliter sized chambers. The fluorescence intensity correlates directly with bacterial growth. This approach demonstrates excellent compatibility with microfluidic design, using minimal reagents while maintaining a strong signal-to-noise ratio. Jusková et al. employed oxygen-sensitive fluorescent nanoprobes to track bacterial respiration in a gas tight microchamber array.70 The fluorescence intensity of these probes varies with dissolved oxygen concentration, providing a real-time, label-free readout of metabolic activity. The assay achieved MIC detection in a short timeframe and was sensitive to cell densities as low as 25–30 CFU per chamber. This method is highly compatible with thermoplastic microfluidic materials and offers continuous monitoring without requiring cell lysis or dye exchange. Chatzimichail et al. introduced a high-resolution platform using 16S rRNA-based multiplexed FISH combined with imaging flow cytometry for simultaneous pathogen identification and phenotypic AST.71 Their adaptive microfluidic channel design facilitates highly efficient bacterial trapping and rapid fluorescent probe hybridization. A convolutional neural network (CNN) is used to interpret subtle phenotypic changes post-antibiotic exposure, enabling rapid susceptibility profiling. The use of RNA-targeted fluorescence allows for specific and highly resolved single-cell analysis, though it currently requires fixed cells and sophisticated image analysis tools. Liu et al. proposed a dual-mode nanomaterial-based detection system incorporating MnO2@ZIF-90 nanoprobes that produce both colorimetric and fluorescence signals in response to bacterial ATP release and metabolic activity.72 When bacteria are active, they reduce the MnO2 shell and trigger ATP-responsive fluorescence, producing quantifiable color and light emission signals within 5 minutes, specific principle shown in Fig. 5A. The platform achieves detection limits as low as 1 CFU mL−1 and supports smartphone-based readout, the platform shows promise for portable and near patient use.
 | ||
| Fig. 5 Representative microfluidic AST optical detection methods. (A) Integrated microfluidic platform for rapid antibiotic susceptibility testing, featuring bacterial isolation, Gram identification, and parallel AST units; utilizes MnO2@ZIF-90 nanoprobes for dual colorimetric and fluorescence detection of pathogenic bacteria in blood, enabling multiplexed, high-throughput analysis with smartphone-based readout. Reproduced with permission from ref. 72. Copyright 2024, Elsevier. (B) Workflow of a laser-based angle-resolved scattering (ARS) microfluidic platform for droplet-based AST. Picoliter droplets are imaged in flow using ARS, and the resulting images are analyzed by a convolutional neural network to predict optical density, enabling determination of bacterial growth and antibiotic susceptibility at the single-droplet level. Reproduced with permission from ref. 77. Copyright 2025, Elsevier. (C) Integrated microfluidic ACGM device for multiplex SERS-based AST, enabling sequential on-chip bacteria loading, automated antibiotic gradient generation, air-based chamber isolation, buffer washing, and final SERS readout in microwells for rapid and high-throughput MIC determination. Reproduced with permission from ref. 82. Copyright 2022, Royal Society of Chemistry. (D) Workflow of the DropDeepL AST platform, which uses a convolutional neural network to rapidly and automatically classify bacterial growth in nanoliter microfluidic droplets based on bright-field images, enabling high-throughput and accurate antibiotic susceptibility testing within 2 hours. Reproduced with permission from ref. 86. Copyright 2024, Elsevier. | ||
Feng et al. constructed a multilayer microfluidic device integrating SYTO9/PI dual staining with morphometric analysis of bacterial elongation.73 This chip allows for side-by-side comparison of bacterial cells under different antibiotic conditions. While SYTO9/PI fluorescence distinguishes live from dead cells, β-lactam-induced elongation serves as an additional phenotypic marker. Image acquisition and processing are completed within 1 hour, yielding results that correlate with gold standard methods at over 98% accuracy. The platform features 12 isolated drug chambers and can process urine samples directly without prior purification.
These diverse applications of fluorescence detection in microfluidic AST platforms highlight the versatility of this signal modality. From metabolic dyes and respiration sensors to genetic probes and nanomaterial-based logic gates, fluorescent markers offer powerful capabilities for accelerating AST workflows. Despite challenges in standardization and device integration, their continued evolution supports the transition of microfluidic AST from laboratory tools to clinically viable diagnostics.
Dixneuf et al. utilized forward/side light scattering and metabolic auto-fluorescence in flow cytometry, combined with principal component analysis and one class support vector machine algorithms, to rapidly distinguish resistant and sensitive strains.76 This method, fully label-free and compatible with continuous-flow microfluidics, offers promising clinical potential. Graf et al. employed angle-resolved light scattering imaging of droplets encapsulating single bacterial cells.77 Using a convolutional neural network (EfficientNetV2-XL), they predicted bacterial growth status and MICs within a relatively short time for E. coli and Staphylococcus aureus. The approach, offering high throughput and no labeling, is well suited for automated droplet-based microfluidic systems (Fig. 5B). Yang et al. developed a dynamic holographic laser speckle imaging technique to assess bacterial motion under antibiotic treatment.78 Analysis with an artificial neural network enabled label-free MIC determination in a short timeframe with a detection limit of 103 CFU mL−1. Its simple optics and rapid output make it suitable for compact, low-cost diagnostic platforms.
Overall, these unmarked optical imaging techniques demonstrate the feasibility of rapid, high-resolution AST without labeling, using light-scattering signatures and advanced computational analysis. Their integration with microfluidic systems offers an efficient path toward robust, miniaturized, and accessible diagnostic platforms capable of providing clinically actionable results within a few hours.
Chen et al. introduced a 3D-ACEK/SERS microfluidic chip that separates bacteria from whole blood and concentrates them onto a SERS-active electrode for AST.81 By utilizing dielectrophoresis and AC electroosmosis, this platform enables rapid detection of bacterial response to antibiotics with high sensitivity and good compatibility with clinical blood samples. Lin et al. developed an antibiotic concentration gradient microfluidic (ACGM) chip integrated with a reusable SERS substrate82 (Fig. 5C). The device generated 792 reaction chambers exposed to varying antibiotic levels, enabling high-throughput AST with Raman spectra acquired after a 3-hour incubation. The system provided semi-quantitative assessments of MIC and bacterial stress responses, offering both spatial resolution and automation potential.
These studies highlight the versatility of SERS-based Raman detection in microfluidic AST. Whether coupled with electric-field enrichment, droplet encapsulation, gradient generation, or static microchambers, SERS enables rapid, label-free, and multiplex phenotyping.
Beyond chamber-based systems, droplet microfluidics has also benefited from AI-powered analysis. Jeong et al. introduced a multiplexed AST platform using color-coded droplets, where custom image-processing algorithms decoded both the identity and concentration of antibiotics while quantifying bacterial proliferation within hundreds of droplets simultaneously.85 Although their method relied on rule-based algorithms rather than deep learning, this automated image analysis substantially increased throughput and minimized human bias in MIC readout. Most recently, Riti et al. reported a “DropDeepL AST” workflow that combines rapid bright-field imaging of nanoliter droplets with a CNN trained on tens of thousands of droplet images.86 This deep learning classifier enabled highly sensitive and specific detection of bacterial growth, not only matching reference BMD results for colistin resistance but also allowing direct AST on minimally processed urine samples, thus demonstrating both clinical versatility and workflow simplicity (Fig. 5D).
These works illustrate how the synergy between microfluidics and AI-driven image analysis empowers rapid, high-throughput, and label-free phenotypic AST, overcoming traditional limitations of manual interpretation, low throughput, and labor-intensive workflows.
3.2 Electrical detection for phenotypic microfluidic AST
As an milestone in this space, Spencer et al. demonstrated impedance-based phenotypic AST by tracking drug-induced dielectric changes in microchannel-confined bacteria, achieving rapid susceptibility calls at scale.87 This work anticipated later single-cell impedance profiling by establishing robust assay geometry and analysis that link frequency-resolved electrical signatures to growth inhibition. Yang et al. utilized a microfluidic static chamber with nano-structures to trap bacteria and monitor resistance changes over time.88 This platform accurately and rapidly distinguished susceptible from resistant strains using only electrical signals which highlights the potential for minimal-instrumentation, low-cost AST. Chang et al. adopted a high-frequency impedance cytometry strategy and developed a D2/D1 kernel density metric for rapid susceptibility classification.89 Their system required only 20 minutes to deliver accurate AST results across clinical Enterobacteriaceae isolates. Chen et al. integrated single-cell impedance profiling with a fully automated readout pipeline90 (the detection principle is shown in Fig. 6A). By measuring high-frequency opacity shifts after brief drug exposure, this approach offers a promising path toward single-cell AST. These impedance-based platforms demonstrate the potential of rapid, label-free AST using microfluidics, particularly for decentralized or resource-limited settings. Future development may focus on improving sensitivity for low-abundance pathogens and integration with automated data analysis tools.
 | ||
| Fig. 6 Representative microfluidic AST electrical detection methods. (A) Overview of a rapid microfluidic impedance-based AST platform, featuring a seven-electrode chip for high-throughput, label-free single-bacterium viability analysis via frequency-resolved electrical measurements, enabling phenotypic detection of antibiotic susceptibility within minutes. Reproduced with permission from ref. 90. Copyright 2023, John Wiley and Sons. (B) Concept and operation of an all-electrical, paper-based AST device, enabling in situ, real-time monitoring of biofilm viability and antibiotic efficacy via extracellular electron transfer (EET) signals, with rapid MIC quantification and mechanism insight for clinical and industrial application.91 Reproduced with permission from ref. 86. Copyright 2023, Royal Society of Chemistry. | ||
In contrast, Rafiee and Choi introduced a 3D paper-based microbial fuel cell for AST of P. aeruginosa biofilms91 (Fig. 6B). This innovative design captures real-time metabolic activity via electrical current generated by electron transfer processes. Though tailored to electrogenic bacteria and biofilm models, its low cost, portability, and reagent-free operation are promising for antimicrobial screening and resistance profiling in resource-limited environments. Domingo-Roca et al. focused on 3D-printed impedance biosensors using hydrogel overlays for rapid, label-free AST.92 Their platform showcased the potential of fully additive manufacturing to fabricate disposable, customizable electrochemical chips. Crane et al. developed a multiplex electrochemical lab-on-a-chip capable of testing seven antibiotics simultaneously against UTI pathogens using redox dye-based voltammetry.93 With integrated hydrogels and screen-printed electrodes, the device enables rapid result delivery using unprocessed urine. Its robust multiplexing capability and reagent stability underline strong translational potential.
Together, these platforms demonstrate the breadth of electrochemical AST from impedance and redox-based detection to self-powered microbial fuel cell systems, each balancing throughput, speed, and operational simplicity. Electrochemical methods hold particular promise for decentralized testing due to their compact hardware requirements and seamless integration with microfluidic form factors.
3.3 Mechanical detection for phenotypic microfluidic AST
Cantilever-based sensors are a class of nanomechanical devices that transduce bacterial activity into measurable physical displacements or vibrations. Zhou et al. demonstrated a fiber-integrated cantilever system employing Fabry–Pérot interferometry to track nanoscale fluctuations of bacteria immobilized on the cantilever surface.94 Antibiotic exposure reduced the vibrational variance significantly, enabling bacterial viability determination. The compact optical design requires no labeling or imaging and is compatible with fluidic environments. Building on this principle, Sturm et al. developed a high-throughput platform using atomic force microscopy cantilevers combined with machine learning to classify nanomotion signals across thousands of clinical isolates.23 Their system accurately predicted susceptibility using blood culture samples, showing strong clinical potential. Taken together, these studies highlight the utility of cantilever-based nanomechanical sensing as a fast, label-free, and growth-independent approach to AST.In addition to cantilever sensors, mechanical detection in microfluidic AST can also leverage deformation-based techniques. Chen et al. presented a simulation-based study integrating cell squeezing and pulsed electric fields to enhance intracellular delivery through transient membrane poration.95 By coupling hydrodynamic stress in narrow microchannels with localized electroporation, they demonstrated significantly improved membrane permeability at lower electric field strengths. While the study is theoretical, it provides a promising framework for mechanical–electrical hybrid platforms capable of manipulating single cells in flow. Such methods may be repurposed for rapid viability assessment and drug susceptibility analysis, especially in hard-to-treat pathogens where delivery of diagnostic agents or stressors is critical.
4. Challenges and opportunities
4.1 Single-cell phenotypic AST
Single-cell phenotypic AST addresses limitations of bulk assays by resolving resistance heterogeneity, shortening detection windows, and helping to deconvolve polymicrobial infections capabilities for which microfluidic methods are particularly well suited. We previously also mentioned that many studies use single-cell detection. Kandavalli et al. developed a microfluidic platform that captures single bacterial cells directly from mixed clinical samples, performs species identification by in situ FISH targeting 16S/23S rRNA, and monitors growth under antibiotic exposure with real-time imaging; in the authors' report, species-specific susceptibility calls were obtained in a relatively short timeframe and successfully discriminated resistant from susceptible strains in mixed cultures.96 For droplet-based systems, Graf et al. encapsulated single cells in picoliter oil-isolated droplets and combined angle-resolved light scattering with a convolutional neural network for growth analysis; this label-free approach distinguished resistant and susceptible E. coli and S. aureus strains in a relatively short timeframe and used droplet throughput to improve detection of rare resistant mutants.77 Li et al. used under-oil open microchambers for single-cell live imaging, reporting MIC estimation within a few hours while maintaining high cell viability an advantage when studying slow-growing organisms.22Single-cell AST can capture early growth/death changes at the individual cell level, shorten result interpretation time, and detect rare drug-resistant subpopulations—thus preventing important variations from being masked by population averages. Additionally, it facilitates tracking the response of individual cells and studying the process of drug resistance development. Moreover, the encapsulation or capture of single cells reduces the required bacterial quantity and reagent consumption. As such, it represents a crucial development direction for future antimicrobial susceptibility testing research.
4.2 Integration of sample-to-result workflows
For critical infections such as bloodstream infections (BSIs), the extremely low abundance of pathogens in clinical samples, combined with high background levels of host cells and plasma proteins, necessitates enrichment or pre-culturing steps to achieve the bacterial concentrations suitable for phenotypic AST. These processes can take 6–24 hours and often constitute the main bottleneck for rapid diagnostics. Recent advances have explored the use of surface-functionalized magnetic beads to selectively capture bacteria, followed by on-chip release and immediate nanoliter-scale cultivation. Notably, a recent publication in Nature reported that the integration of magnetic enrichment and microfluidic technology enables a substantial reduction in pre-culture time.97 This study addressed the dominant clinical bottleneck—blood culture—by bypassing culture altogether and recovering pathogens directly from whole blood using synthetic β2-glycoprotein I (sβ2GPI) peptide-coated magnetic nanoparticles, followed by species ID (QmapID) and low-inoculum microfluidic AST on a 96-well-style chip. In a clinical enrollment of 190 suspected-infection patients, the platform achieved 100% species-ID match, and in six retrospectively tested positive cases, AST showed 94.9% categorical agreement with an average theoretical TAT of 13 ± 2.53 h from initial blood processing representing a >40–60 h reduction versus hospital workflows.To further realize seamless workflows, future work should focus on improving capture universality across diverse Gram types and strains, designing automated microfluidic systems to precisely couple release with microchamber inoculation, and ensuring single-use, fully enclosed fluidic paths to prevent cross-contamination.
4.3 Detection of slow-growing and persistent pathogens
Some pathogens, including Mycobacterium tuberculosis (TB), anaerobes, and biofilm-forming bacteria, exhibit prolonged replication cycles and stress tolerance, posing significant challenges for conventional AST approaches reliant on growth-based readouts. Several promising solutions have emerged:Metabolic amplification: use of redox dyes such as resazurin, isotopic labeling with deuterium oxide, or microcalorimetry can detect metabolic activity in low-copy bacterial populations without relying on visible growth.70,98
Nanomechanical single-cell sensing: AFM-based cantilevers or piezoelectric devices can detect subcellular mechanical fluctuations,99 enabling AST in slow growers within 2–4 hours, as shown in recent work.23
Multiphysical stimulation: combining electric fields, acoustic waves, and pressure gradients may enhance nutrient diffusion and reduce the lag phase for metabolically dormant pathogens, accelerating the onset of measurable activity.
4.4 Interference from residual antibiotics
Empirical antibiotic administration prior to sampling can leave residual drug concentrations in blood or urine, leading to false-negative AST results or overestimation of resistance. Addressing this requires targeted sample preprocessing strategies:Selective adsorption or enzymatic neutralization: incorporating modules with β-lactamase-coated beads or hydrophobic adsorption surfaces in the sample inlet region to degrade or capture residual antibiotics.100
Dilution and resuspension: following high-speed magnetic pathogen separation, the supernatant can be discarded, and bacteria resuspended in fresh isosmotic media to reduce antibiotic concentrations by one to two orders of magnitude.
Internal controls: including drug-free reference chambers within the same chip allows for intra-device growth comparisons, helping to normalize results against any residual drug effects.
Collectively, these strategies are critical for improving the clinical accuracy and robustness of phenotypic microfluidic AST systems under real-world conditions.
4.5 Multimodal validation and algorithmic integration
Microfluidic AST platforms that rely on a single culture geometry or a single sensing modality may be susceptible to signal ambiguities or model-specific limitations. Incorporating cross-validation strategies, such as embedding both static and flow-through microchambers with parallel readouts including fluorescence imaging and impedance spectroscopy, can improve diagnostic reliability.101,102 Additionally, computational tools are enabling deeper insight and interpretability: machine learning techniques can integrate temporal fluorescence images, impedance spectra, and pressure profiles to build multiparametric classifiers that better predict antibiotic efficacy.Digital twin simulations can model drug diffusion, metabolic byproduct accumulation, and signal transduction dynamics to optimize microfluidic design and reduce experimental trial-and-error.
4.6 Toward standardization
It is worth noting that the above studies differ in the types of bacteria tested, the antibiotics used, and the scope of time measurement—some focus on the total duration of the entire process, while others primarily target the time required for laboratory cultivation and AST itself. As a result, the antimicrobial susceptibility testing times reported across these studies are not directly comparable due to variations in experimental design and reporting metrics. Therefore, further testing under standardized condition with a good coverage of bacteria species can be necessary.For phenotypic microfluidic AST to achieve practical implementation, methodological standardization to ensure consistency with CLSI/EUCAST reference endpoints, facilitating regulatory comparison and clinical acceptance. Regulatory compliance and quality systems for reagent integration, microfluidic actuation (e.g., pumps or valves), and optoelectronic/electrochemical readout modules need to be validated to meet IVD certification. Open-source data repositories that enable sharing of raw multimodal datasets to improve reproducibility, support inter-laboratory benchmarking, and facilitate algorithm generalization across platforms. Addressing these aspects will be critical to transitioning microfluidic AST from promising prototypes to standardized and accessible diagnostic solutions.
Conclusions
Microfluidic technologies have rapidly advanced phenotypic AST, demonstrating unprecedented improvements in assay speed, miniaturization, and automation. Through innovation in cultivation structures and detection strategies, these platforms achieve reliable, high-throughput, and information-rich readouts, sometimes even at the single-cell level. Recent systems integrate sophisticated gradient generation, precise compartmentalization, and advanced data analytics, including AI-driven interpretation, to deliver accurate susceptibility results within just a few hours. Nonetheless, several critical translational challenges remain, including automated sample enrichment from complex clinical matrices, ensuring robust performance with slow-growing or biofilm-forming pathogens, and mitigation of residual antibiotic interference. Future directions should emphasize full sample-to-answer integration, scalable manufacturing, regulatory standardization, and clinical validation. The convergence of microfluidic engineering, materials science, and computational analysis is poised to accelerate the clinical deployment of phenotypic AST. As these hurdles are addressed, microfluidic AST platforms have the potential to revolutionize antimicrobial stewardship, improve patient outcomes, and provide a critical weapon in the ongoing fight against antimicrobial resistance.Author contributions
Mo Shen led conceptualization, writing and analyses. Qi Wang explored and discussed future research directions. Qingqing Luo conducted the statistical analysis of the current development status and completed the proofreading of the manuscript. Jiatong Zhao completed the proofreading of the manuscript. Feng Shen assisted with analyses, writing and editing.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by the Natural Science Foundation of Shanghai (No. 23ZR1432900), the Interdisciplinary Program of Shanghai Jiao Tong University (No. YG2024QNA45, YG2025ZD16), the National Natural Science Foundation of China (No. 32470086), the National Natural Science Foundation of China (No. 32171463).References
- C. J. L. Murray, K. S. Ikuta, F. Sharara and L. Swetschinski, et al. , Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- M. Naghavi, S. E. Vollset and K. S. e. a. Ikuta, Lancet, 2024, 404, 1199–1226 CrossRef PubMed.
- M. A. Salam, M. Y. Al-Amin, J. S. Pawar, N. Akhter and I. B. Lucy, Saudi J. Biol. Sci., 2023, 30, 103582 CrossRef CAS PubMed.
- S. Hattab, A. H. Ma, Z. Tariq, I. Vega Prado, I. Drobish, R. Lee and R. Yee, Antibiotics, 2024, 13, 786 CrossRef CAS PubMed.
- M. Benkova, O. Soukup and J. Marek, J. Appl. Microbiol., 2020, 129, 806–822 CrossRef CAS.
- J. O'Neill, Tackling drug-resistant infections globally: final report and recommendations, The Review on Antimicrobial Resistance, HM Government and Wellcome Trust, 2016 Search PubMed.
- Ö. Baltekin, A. Boucharin, E. Tano, D. I. Andersson and J. Elf, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9170–9175 CrossRef.
- D. K. Sahel, G. Giriprasad, R. Jatyan, S. Guha, A. Korde, A. Mittal, S. Bhand and D. Chitkara, RSC Adv., 2024, 14, 32411–32435 RSC.
- J. Choi, Y.-G. Jung, J. Kim, S. Kim, Y. Jung, H. Na and S. Kwon, Lab Chip, 2013, 13, 280–287 RSC.
- S. H. Needs, J. Pivetal, J. Hayward, S. P. Kidd, H. Lam, T. Diep, K. Gill, M. Woodward, N. M. Reis and A. D. Edwards, Sens. Diagn., 2023, 2, 736–750 RSC.
- W. Postek and P. Garstecki, Acc. Chem. Res., 2022, 55, 605–615 CrossRef CAS.
- G. Reszetnik, K. Hammond, S. Mahshid, T. AbdElFatah, D. Nguyen, R. Corsini, C. Caya, J. Papenburg, M. P. Cheng and C. P. Yansouni, Nat. Commun., 2024, 15, 9719 CrossRef CAS.
- W. Postek, N. Pacocha and P. Garstecki, Lab Chip, 2022, 22, 3637–3662 RSC.
- M. Zhu, T. Xu, Y. Cheng, B. Ma, J. Xu, Z. Diao, F. Wu, J. Dai, X. Han, P. Zhu, C. Pang, J. Li, H. Wang, R. Xu and X. Li, Anal. Chem., 2023, 95, 14375–14383 CrossRef CAS.
- W. Wu and Y. Mu, Biomicrofluidics, 2024, 18, 031504 CrossRef CAS PubMed.
- A. V. Nguyen, M. Yaghoobi, M. Azizi, M. Davaritouchaee, K. W. Simpson and A. Abbaspourrad, Commun. Eng., 2023, 2, 15 CrossRef CAS.
- Q. Wang, X. Li, Y. a. Ren, Q. Hu, L. Xu, W. Chen, J. Liu, N. Wu, M. Tao, J. Sun, Y. Xu and F. Shen, Biosens. Bioelectron., 2025, 271, 117084 CrossRef CAS.
- W. Lu, H. Li, H. Qiu, L. Wang, J. Feng and Y. V. Fu, Front. Microbiol., 2023, 13, 127907 Search PubMed.
- Z. Diao, A. Ge, H. Zhou, X. Zhou, L. Kan, W. Gao, W. Shen, Y. Ji, H. Wang, J. Xu, X. Wang and B. Ma, Sens. Actuators, B, 2025, 442, 138183 CrossRef.
- J. Chen, J. Zhong, Y. Chang, Y. Zhou, S. H. Koo, T. Y. Tan, H. Lei and Y. Ai, Small, 2024, 20, 2303352 CrossRef CAS.
- M. S. Chiriacò, E. Primiceri, F. De Feo, A. Montanaro, A. G. Monteduro, A. Tinelli, M. Megha, D. Carati and G. Maruccio, Biosens. Bioelectron., 2016, 79, 9–14 CrossRef CAS.
- C. Li, S. McCrone, J. W. Warrick, D. R. Andes, Z. Hite, C. F. Volk, W. E. Rose and D. J. Beebe, Lab Chip, 2023, 23, 2005–2015 RSC.
- A. Sturm, G. Jóźwiak, M. P. Verge, L. Munch, G. Cathomen, A. Vocat, A. Luraschi-Eggemann, C. Orlando, K. Fromm, E. Delarze, M. Świątkowski, G. Wielgoszewski, R. M. Totu, M. García-Castillo, A. Delfino, F. Tagini, S. Kasas, C. Lass-Flörl, R. Gstir, R. Cantón, G. Greub and D. Cichocka, Nat. Commun., 2024, 15, 2037 CrossRef CAS PubMed.
- P. Zhang, A. M. Kaushik, K. Hsieh, S. Li, S. Lewis, K. E. Mach, J. C. Liao, K. C. Carroll and T.-H. Wang, Small Methods, 2022, 6, 2101254 CrossRef CAS PubMed.
- Y.-T. Kao, T. S. Kaminski, W. Postek, J. Guzowski, K. Makuch, A. Ruszczak, F. von Stetten, R. Zengerle and P. Garstecki, Lab Chip, 2020, 20, 54–63 RSC.
- Y. Z. Ahmed Nawaz Qureshi, M. Li, H. Chang and Y. Song, Biosens. Bioelectron., 2025, 273, 117160 CrossRef CAS.
- J. S. Kim, J. Kim, J.-S. Kim, W. Kim and C.-S. Lee, Lab Chip, 2024, 24, 5274–5289 RSC.
- G.-S. Du, J.-Z. Pan, S.-P. Zhao, Y. Zhu, J. M. J. den Toonder and Q. Fang, Anal. Chem., 2013, 85, 6740–6747 CrossRef CAS PubMed.
- P. Sun, Y. Liu, J. Sha, Z. Zhang, Q. Tu, P. Chen and J. Wang, Biosens. Bioelectron., 2011, 26, 1993–1999 CrossRef CAS PubMed.
- G. S. Ugolini, M. Wang, E. Secchi, R. Pioli, M. Ackermann and R. Stocker, Lab Chip, 2024, 24, 1394–1418 RSC.
- T. Xu, X. Han, P. Zhu, J. Dai, M. Liu, Y. Liu, J. Xu and B. Ma, Sens. Actuators, B, 2021, 343, 130117 CrossRef CAS.
- E. Grigorov, S. Peykov and B. Kirov, Appl. Sci., 2022, 12, 2198 CrossRef CAS.
- A. V. Nguyen, M. Azizi, M. Yaghoobi, B. Dogan, S. Zhang, K. W. Simpson and A. Abbaspourrad, Anal. Chem., 2021, 93, 5789–5796 CrossRef CAS.
- L. Ma, M. Petersen and X. Lu, Appl. Environ. Microbiol., 2020, 86, e00096-20 CrossRef.
- R. Wang and D. Erickson, ACS Omega, 2021, 6, 1410–1414 CrossRef CAS.
- M. Azizi, B. Davaji, A. V. Nguyen, S. Zhang, B. Dogan, K. W. Simpson and A. Abbaspourrad, ACS Sens., 2021, 6, 1560–1571 CrossRef CAS.
- Z. Pang, S. Li, S. Wang, Z. Cai, S. Zhang, C. Wan, J. Wang, Y. Li, P. Chen and B.-F. Liu, Anal. Chim. Acta, 2024, 1287, 342033 CrossRef CAS.
- F. Shen, W. Du, J. E. Kreutz, A. Fok and R. F. Ismagilov, Lab Chip, 2010, 10, 2666–2672 RSC.
- F. Shen, in Microchip Diagnostics: Methods and Protocols, ed. V. Taly, J.-L. Viovy and S. Descroix, Springer New York, New York, NY, 2017, pp. 123–132, DOI:10.1007/978-1-4939-6734-6_10.
- F. Shen, W. Du, E. K. Davydova, M. A. Karymov, J. Pandey and R. F. Ismagilov, Anal. Chem., 2010, 82, 4606–4612 CrossRef CAS.
- Q. Yi, D. Cai, M. Xiao, M. Nie, Q. Cui, J. Cheng, C. Li, J. Feng, G. Urban, Y.-C. Xu, Y. Lan and W. Du, Biosens. Bioelectron., 2019, 135, 200–207 CrossRef CAS PubMed.
- G. Cai, W. Wu, S. Feng and Y. Liu, Analyst, 2021, 146, 4622–4629 RSC.
- X. Li, X. Liu, Z. Yu, Y. Luo, Q. Hu, Z. Xu, J. Dai, N. Wu and F. Shen, Lab Chip, 2022, 22, 3952–3960 RSC.
- X. Liu, X. Li, N. Wu, Y. Luo, J. Zhang, Z. Yu and F. Shen, ACS Sens., 2022, 7, 1977–1984 CrossRef CAS PubMed.
- S. Kim, S. Lee, J.-K. Kim, H. J. Chung and J. S. Jeon, Biomicrofluidics, 2019, 13, 014108 CrossRef PubMed.
- X. Cui, L. M. Lee, X. Heng, W. Zhong, P. W. Sternberg, D. Psaltis and C. Yang, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10670–10675 CrossRef CAS PubMed.
- Z. Long, E. Nugent, A. Javer, P. Cicuta, B. Sclavi, M. Cosentino Lagomarsino and K. D. Dorfman, Lab Chip, 2013, 13, 947–954 RSC.
- P. Wistrand-Yuen, C. Malmberg, N. Fatsis-Kavalopoulos, M. Lübke, T. Tängdén, J. Kreuger and J. M. Bomberger, mBio, 2020, 11, 3109–3119 CrossRef.
- N. Blanco-Cabra, M. J. López-Martínez, B. V. Arévalo-Jaimes, M. T. Martin-Gómez, J. Samitier and E. Torrents, npj Biofilms Microbiomes, 2021, 7, 62 CrossRef CAS PubMed.
- H. Jeon, Z. A. Khan, E. Barakat and S. Park, Antibiotics, 2020, 9, 348 CrossRef CAS PubMed.
- Z. Pang, L. Shi, M. Wang and J. Tao, Analyst, 2025, 150, 1398–1408 RSC.
- Z. Pang, L. Shi, Y. Chai, L. Wan, X. Zhang, M. Wang and J. Tao, Microchem. J., 2025, 209, 112685 CrossRef CAS.
- G. Amselem, S. Sart and C. N. Baroud, in Methods in Cell Biology, ed. D. A. Fletcher, J. Doh and M. Piel, Academic Press, 2018, vol. 148, pp. 177–199 Search PubMed.
- M. N. Hatori, S. C. Kim and A. R. Abate, Anal. Chem., 2018, 90, 9813–9820 CrossRef CAS PubMed.
- P. Sabhachandani, S. Sarkar, P. C. Zucchi, B. A. Whitfield, J. E. Kirby, E. B. Hirsch and T. Konry, Microchim. Acta, 2017, 184, 4619–4628 CrossRef CAS.
- M. Azizi, A. V. Nguyen, B. Dogan, S. Zhang, K. W. Simpson and A. Abbaspourrad, ACS Appl. Mater. Interfaces, 2021, 13, 19581–19592 CrossRef CAS.
- A. Vasala, V. P. Hytönen and O. H. Laitinen, Front. Cell. Infect. Microbiol., 2020, 10, e00308 Search PubMed.
- B. Behera, G. K. Anil Vishnu, S. Chatterjee, V. S. N. Sitaramgupta V, N. Sreekumar, A. Nagabhushan, N. Rajendran, B. H. Prathik and H. J. Pandya, Biosens. Bioelectron., 2019, 142, 111552 CrossRef CAS.
- M. L. Kovarik, D. M. Ornoff, A. T. Melvin, N. C. Dobes, Y. Wang, A. J. Dickinson, P. C. Gach, P. K. Shah and N. L. Allbritton, Anal. Chem., 2013, 85, 451–472 CrossRef CAS PubMed.
- P. S. Dittrich and A. Manz, Nat. Rev. Drug Discovery, 2006, 5, 210–218 CrossRef CAS.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS.
- J. Zhang, C. Ma, Y. Du, J. Huang and L. Xue, Front. Chem., 2025, 13, 1536928 CrossRef CAS PubMed.
- A. Kulesa, J. Kehe, J. E. Hurtado, P. Tawde and P. C. Blainey, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 6685–6690 CrossRef PubMed.
- X. Liu, R. E. Painter, K. Enesa, D. Holmes, G. Whyte, C. G. Garlisi, F. J. Monsma, M. Rehak, F. F. Craig and C. A. Smith, Lab Chip, 2016, 16, 1636–1643 RSC.
- D. I. Njoku, Q. Guo, W. Dai, J. L. Chen, G. Mao, Q. Sun, H. Sun and Y.-K. Peng, TrAC, Trends Anal. Chem., 2023, 167, 117288 CrossRef CAS.
- T. Dong and X. Zhao, Anal. Chem., 2015, 87, 2410–2418 CrossRef CAS PubMed.
- W. Wu, G. Cai, Y. Liu, Y. Suo, B. Zhang, W. Jin, Y. Yu and Y. Mu, Lab Chip, 2023, 23, 2399–2410 RSC.
- J. C. Palomino, A. Martin, M. Camacho, H. Guerra, J. Swings and F. Portaels, Antimicrob. Agents Chemother., 2002, 46, 2720–2722 CrossRef CAS.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421–5426 CrossRef.
- P. Jusková, S. Schmitt, A. Kling, D. G. Rackus, M. Held, A. Egli and P. S. Dittrich, ACS Sens., 2021, 6, 2202–2210 CrossRef PubMed.
- S. Chatzimichail, P. Turner, C. Feehily, A. Farrar, D. Crook, M. Andersson, S. Oakley, L. Barrett, H. El Sayyed, J. Kyropoulos, C. Nellåker, N. Stoesser and A. N. Kapanidis, Lab Chip, 2024, 24, 4843–4858 RSC.
- X. Liu, X. Chen, S. Yin, Y. Liu, W. Shi, F. Wang and C. Li, Chem. Eng. J., 2024, 499, 156276 CrossRef.
- L. Feng, T. Zhang, H. Tang, J. Zhang, X. Ma, J. Pang, L. Chen, W. Wang, Y. Wang, H. Wu and G. Zhou, Talanta, 2025, 295, 128388 CrossRef CAS PubMed.
- B. Péter, E. Farkas, S. Kurunczi, Z. Szittner, S. Bősze, J. J. Ramsden, I. Szekacs and R. Horvath, Biosensors, 2022, 12, 188 CrossRef PubMed.
- P. Wang, H. Sun, W. Yang and Y. Fang, Biosensors, 2022, 12, 1171 CrossRef CAS.
- S. Dixneuf, A.-C. Chareire-Kleiberg, P. Mahé, M. El Azami, C. Kolytcheff, S. Bellais, C. Guyard, C. Védrine, F. Mallard, Q. Josso and F. Rol, Front. Microbiol., 2023, 14, 1232250 CrossRef PubMed.
- M. Graf, A. Sarkar, C.-M. Svensson, A.-S. Munser, S. Schröder, S. Hengoju, M. A. Rosenbaum and M. T. Figge, Sens. Actuators, B, 2025, 424, 136866 CrossRef CAS.
- J. Yang, K. Zhou, C. Zhou, P. S. Khamsi, O. Voloshchuk, L. Hernandez, J. Kovac, A. Ebrahimi and Z. Liu, Biosens. Bioelectron., 2025, 278, 3462 Search PubMed.
- N. E. Dina, M. A. Tahir, S. Z. Bajwa, I. Amin, V. K. Valev and L. Zhang, Biosens. Bioelectron., 2023, 219, 114843 CrossRef CAS PubMed.
- A. Fraiman and L. D. Ziegler, Talanta, 2025, 292, 114843 CrossRef PubMed.
- K.-H. Chen, S.-H. Lee, L.-C. Kok, T.-O. Ishdorj, H.-Y. Chang and F.-G. Tseng, Biosens. Bioelectron., 2022, 197, 113740 CrossRef CAS.
- S.-J. Lin, P.-H. Chao, H.-W. Cheng, J.-K. Wang, Y.-L. Wang, Y.-Y. Han and N.-T. Huang, Lab Chip, 2022, 22, 1805–1814 RSC.
- H. Yu, W. Jing, R. Iriya, Y. Yang, K. Syal, M. Mo, T. E. Grys, S. E. Haydel, S. Wang and N. Tao, Anal. Chem., 2018, 90, 6314–6322 CrossRef CAS PubMed.
- A. A. Sklavounos, C. R. Nemr, S. O. Kelley and A. R. Wheeler, Lab Chip, 2021, 21, 4208–4222 RSC.
- Y. Jeong, H. Jang, J. Kang, J. Nam, K. Shin, S. Kwon and J. Choi, Biosensors, 2021, 11, 283 CrossRef CAS PubMed.
- J. Riti, G. Sutra, T. Naas, H. Volland, S. Simon and K. Perez-Toralla, Biosens. Bioelectron., 2024, 257, 116301 CrossRef CAS PubMed.
- D. C. Spencer, T. F. Paton, K. T. Mulroney, T. J. J. Inglis, J. M. Sutton and H. Morgan, Nat. Commun., 2020, 11, 5328 CrossRef CAS.
- Y. Yang, K. Gupta and K. L. Ekinci, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10639–10644 CrossRef.
- Y. Chang, J. Zhong, A. K. Bin Kassim, S. H. Koo, T. Y. Tan, Y. Zhou and Y. Ai, Sens. Actuators, B, 2025, 423, 116301 CrossRef.
- J. Chen, J. Zhong, Y. Chang, Y. Zhou, S. H. Koo, T. Y. Tan, H. Lei and Y. Ai, Small, 2023, 20, 2303352 CrossRef.
- Z. Rafiee and S. Choi, Analyst, 2023, 148, 2501–2510 RSC.
- R. Domingo-Roca, P. Lasserre, L. Riordan, A. R. Macdonald, A. Dobrea, K. R. Duncan, S. Hannah, M. Murphy, P. A. Hoskisson and D. K. Corrigan, Biosens. Bioelectron.: X, 2023, 13, 100308 CAS.
- B. Crane, A. Iles, C. E. Banks, M. Rashid, P. E. Linton and K. J. Shaw, Biomed. Microdevices, 2024, 26, 35 CrossRef CAS.
- J. Zhou, J. Huang, H. Huang, C. Zhao, M. Zou, D. Liu, X. Weng, L. Liu, J. Qu, L. Liu, C. Liao and Y. Wang, Biomed. Opt. Express, 2023, 14, 1862–1873 CrossRef CAS.
- J. Chen, H. Liu, C. Li, X. Chen and Y. Dai, Microfluid. Nanofluid., 2023, 28, 4 CrossRef.
- V. Kandavalli, P. Karempudi, J. Larsson and J. Elf, Nat. Commun., 2022, 13, 6215 CrossRef CAS PubMed.
- T. H. Kim, J. Kang, H. Jang, H. Joo, G. Y. Lee, H. Kim, U. Cho, H. Bang, J. Jang, S. Han, D. Y. Kim, C. M. Lee, C. K. Kang, P. G. Choe, N. J. Kim, M.-d. Oh, T. S. Kim, I. Kim, W. B. Park and S. Kwon, Nature, 2024, 632, 893–902 CrossRef CAS PubMed.
- Y. Liu, T. Lehnert, T. Mayr and M. A. M. Gijs, Lab Chip, 2021, 21, 3520–3531 RSC.
- A. Vocat, A. Sturm, G. Jóźwiak, G. Cathomen, M. Świątkowski, R. Buga, G. Wielgoszewski, D. Cichocka, G. Greub and O. Opota, Microbes Infect., 2023, 25, 105151 CrossRef CAS PubMed.
- Z. Su, W. Hu, L. Ye, D. Gao and J.-M. Lin, Chin. Chem. Lett., 2023, 34, 107790 CrossRef CAS.
- D. Zhang, Y. Zhang, F. Yin, Q. Qin, H. Bi, B. Liu and L. Qiao, Analyst, 2021, 146, 515–520 RSC.
- W. Lu, H. Li, H. Qiu, L. Wang, J. Feng and Y. V. Fu, Front. Microbiol., 2023, 13, 1076965 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |
