Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Programmable self-destruction of artificial cells with death signaling
Joshua
Krehan
,
Lorena
Baranda Pellejero
and
Andreas
Walther†
 *
*
Life-Like Materials and Systems, Department of Chemistry, University of Mainz, Duesbergweg 10-14, 55128 Mainz, Germany. E-mail: Andreas.Walther@uni-mainz.de
First published on 8th December 2025
Abstract
Programmed cell death is a crucial biological process that removes damaged or no longer needed cells and is orchestrated through complex intracellular signaling cascades. Mimicking such behavior in synthetic systems enables programmed disassembly after completing a task or releasing cargo on demand. Despite advances in engineering artificial cells (ACs) that mimic key cellular functions such as metabolism, homeostasis or communication, systems with a programmable lifetime remain unrealized. Here, we introduce a time-programmed self-destruction mechanism in ACs based on pH-responsive giant unilamellar vesicles, equipped with an internal UV-inducible acidification cascade. Upon light activation, photocaged glucose is enzymatically converted to gluconic acid, lowering the internal pH and destabilizing the pH-sensitive membrane, ultimately causing complete membrane collapse. The self-destruction is spatially confined and tunable in time, ranging from minutes to over an hour, depending on the light intensity. Furthermore, we demonstrate that collapse-induced release of DNA signals triggers defined downstream responses in neighboring ACs, including membrane labeling and aggregation. Our findings pave the way for ACs with programmed lifetimes, capable of on-demand release or disassembly in response to defined stimuli, allowing transmission of signals within complex synthetic environments.
Introduction
Programmed cell death is a tightly regulated biological process through which multicellular organisms remove damaged or unnecessary cells in response to internal or external signals.1–3 These signals trigger intracellular biochemical cascade reactions that lead to the controlled disassembly of cells and enable the release of molecular messengers, which then often activate downstream immune responses or regenerative programs in neighboring cells.4–6 Mimicking such behavior in synthetic systems holds potential for engineering life-like materials with autonomous disassembly, controlled lifetimes, or programmable communication. Artificial cells (ACs) have been engineered to replicate key biological functions such as controlled growth and division,7–10 response to their environment,11–14 and basic intercellular communication.15–18 More complex communication has further enabled programmable signal transport19 and DNA-based catalytic networks for chemical adaptation,20 and has recently been extended to AC-based spheroids in which inter-AC communication and homeostasis emerge as collective properties.21 While external light-induced bursting of lipid vesicles has been demonstrated using photosensitive surfactants,22 ACs that undergo a programmed, cell death-like disassembly via an internal mechanism, resulting in complete collapse, have not yet been achieved.Enzymatic pH-feedback mechanisms enable autonomous pH control by coupling substrate consumption with the pH dependency of enzyme activity.23 Two prominent examples of pH-modulating enzymes are urease, hydrolyzing urea to ammonia and carbon dioxide and thereby increasing the pH, and glucose oxidase (GOx), converting glucose into gluconic acid and thereby lowering the pH.24–26 Beyond operation in plain solution, such enzymes have been compartmentalized in hydrogels,27,28 polymer beads,29 polymer capsules,21 liposomes,30–32 and polymersomes,33–38 where they effectively adjust their surrounding pH while maintaining stability of the compartments. To date, no mechanism has been established that triggers pH modulation strictly from within a compartment, which is related to the challenge of keeping a system dormant during assembly and enabling external triggering at defined times. To this end, photochemical uncaging of substrates for enzymes can provide an important spatiotemporal control mechanism.39–41
Herein, we introduce a time-programmed AC kill switch that drives the controlled self-destruction of ACs based on internal, UV-inducible pH modulation coupled to a pH-responsive giant unilamellar vesicle-based (GUV) chassis. Importantly, our design ensures that ACs remain stable until the self-destruction mechanism is activated by UV irradiation, after which they undergo irreversible collapse. The timing of collapse can be programmed from minutes to hours by tuning the UV intensity, and the response is spatially confined such that only ACs within the irradiated area self-destruct. We further demonstrate downstream signaling by showing that DNA released after self-destruction can be sensed on the membrane of intact receiver ACs and can even drive aggregation of ACs.
Results
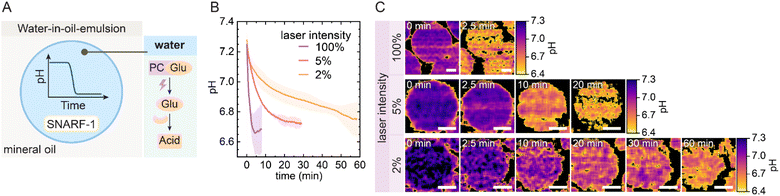
Our system design relies on pH-responsive GUV-based ACs encapsulating a UV-inducible acidification cascade by co-loading a photocaged glucose derivative (PCGlu)42,43 together with GOx (Fig. 1A). Upon UV irradiation, glucose is released from its photocaged form and subsequently converted by GOx to generate glucono-1,5-lactone, which rapidly hydrolyzes into gluconic acid, resulting in a gradual internal acidification. The GUV membrane is engineered with a lipid mixture consisting of DOPC (neutral fluid lipid), DODAP (cationic lipid), and CHEMS (cholesteryl hemisuccinate, anionic pH-sensitive lipid) (Fig. 1B).44 At neutral pH, the membrane is stable due to the electrostatic balance between the lipids. However, once the internal pH (pHi) drops below a threshold at slightly acidic pH, protonation of CHEMS disrupts the charge balance of the membrane, triggering destabilization and collapse with complete release of the encapsulated content.We first demonstrate the UV-triggered, temporally programmable acidification of the PCGlu-GOx system in a simplified model using water-in-oil emulsions under conditions that mimic the internal environment of ACs (Fig. 2A). To monitor pH changes upon UV activation, we used 10 mM PCGlu, 1 g per L GOx, and 3 µM SNARF-1-dextran in PBS-buffered droplets (phosphate-buffered saline, 10 mM, pH 7.3). SNARF-1 is a ratiometric fluorescent pH indicator compatible with confocal laser scanning microscopy (CLSM). This enables imaging of individual water droplets or ACs and simultaneous pH measurement. During the experiment, the 405 nm laser of the CLSM continuously uncages PCGlu to release glucose. The kinetics of acidification depend directly on laser intensity: at 100%, the pH drops from ∼7.2 to ∼6.7 within 2 min, while 5% and 2% intensities result in the same final pH after ∼30 min and ∼60 min, respectively (Fig. 2B). Fluorescence-based pH maps of individual droplets confirm these trends and visualize the temporal progression of acidification (Fig. 2C). Together, these results demonstrate that the UV-triggered PCGlu-GOx system enables a tunable pH drop in confined compartments, with acidification timescales adjustable from minutes to over an hour depending on laser intensity.
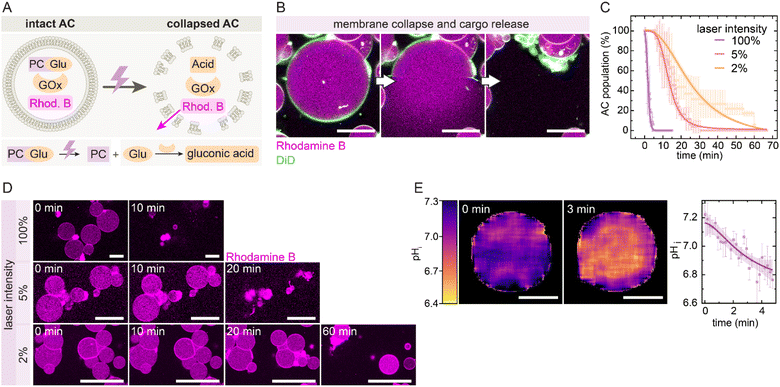
We then applied this system in GUV-based ACs, prepared by the phase transfer method,45,46 to demonstrate UV-triggered self-destruction and cargo release (Fig. 3A). ACs were loaded with 10 mM PCGlu, 1 g per L GOx, and 30 µM of the fluorescent cargo Rhodamine B, which enables visualization of both membrane integrity and release behavior. Upon UV activation, the internal reaction lowers the pHi, resulting in leakage of cargo and complete membrane collapse (Fig. 3B). Time-lapse CLSM confirms that the fraction of intact ACs decreases over time as a function of the laser intensity (Fig. 3C and D, SI Movie 1). At 100% laser intensity, most ACs collapse within 5 min, while 5% and 2% intensities lead to delayed collapse over 30–60 min. The collapse correlates with a measurable drop in pHi, as calculated from the encapsulated SNARF-1 dye (Fig. 3E). Notably, the pHi decreases from ∼7.2 to ∼6.8 within 3 min at 100% intensity and thus reaches the critical range for CHEMS protonation and membrane destabilization. Control experiments confirm that the collapse mechanism is driven by internal acidification and depends on the pH-responsive lipid composition, as well as on parameters of the internal acidification cascade: no self-destruction occurs without GOx (SI Fig. S1A) or with non-pH-responsive DOPC membranes (SI Fig. S1B). Importantly, the generated H2O2 does not induce critical membrane oxidation or collapse, as further evidenced by the stability of DOPC ACs containing PCGlu and GOx. Note that H2O2 is a poor oxidant for lipid oxidation in the absence of catalysts.47,48 Low enzyme loading (≤0.001 g per L GOx, SI Fig. S1C), limited substrate concentration (≤4 mM PCGlu, SI Fig. S1D), or low CHEMS content (≤5% CHEMS, SI Fig. S1E) prevent collapse, while excessive CHEMS results in low vesicle yield during fabrication and predominantly permeable ACs (≥15% CHEMS, SI Fig. S1F). Moreover, the UV-triggered collapse can be spatially confined: by irradiating a defined region, only ACs within that area collapse, while neighboring ACs remain intact (SI Fig. S2). This underscores that a supercritical local acidification triggers the collapse, whereas its further dilution into the surrounding medium prevents the destruction of neighboring ACs. Together, these results confirm that UV-triggered acidification provides a reliable and spatially addressable mechanism to trigger time-programmed AC self-destruction.
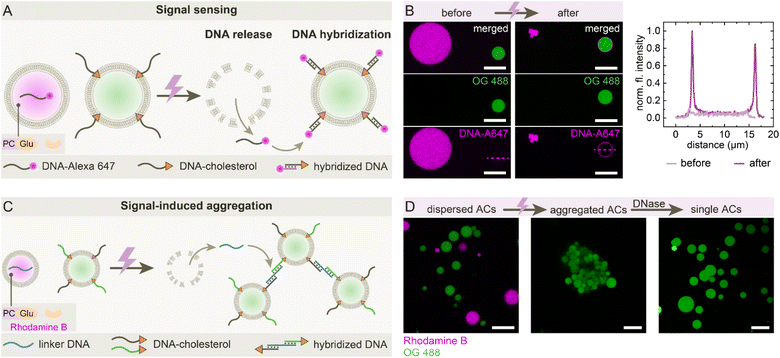
Finally, we designed a system in which UV-triggered self-destruction of sender ACs leads to signal release and downstream responses in neighboring receiver ACs, which do not feature the self-destruction mechanism (Fig. 4A). Sender ACs encapsulate the PCGlu-GOx system and a single-stranded DNA labeled with Alexa 647 (DNA-A647), while receiver ACs are functionalized with membrane-anchored complementary DNA-cholesterol. Upon UV activation, the sender ACs self-destruct and release DNA-A647 into the local environment. The released DNA-A647 then hybridizes with its complementary strand on the receiver AC membrane, resulting in localized membrane fluorescence (Fig. 4B). Line profile analysis confirms this transition. Building on this signaling, we implemented a second response module involving a linker DNA to mediate signal-induced aggregation (Fig. 4C). Upon UV activation, the released DNA bridges receiver ACs via hybridization with DNA-cholesterol on their surfaces, leading to visible AC clustering (Fig. 4D, further overview images of clustering in SI Fig. S3). The aggregation can be reversed with DNase treatment, confirming the DNA-based mechanism. Together, these findings demonstrate that ACs can be equipped with communication modules based on self-destruction, capable of releasing molecular signals to elicit specific death responses in their environment.
Conclusion
In summary, we have introduced a temporally programmable cell death-like AC self-destruction mechanism based on an internally triggered, light-inducible acidification cascade that enables the irreversible collapse of specifically engineered GUV-based ACs. By combining photocaged glucose and glucose oxidase with a pH-sensitive lipid membrane, we achieved a controlled and tunable drop in internal pH, resulting in membrane destabilization and complete AC disassembly. The collapse process is irreversible, spatially confined, and can be precisely timed from minutes to over an hour by adjusting the light intensity. Beyond that, we demonstrated that AC self-destruction can be coupled to downstream signaling, including membrane-localized signal sensing and aggregation in neighboring intact ACs, thereby establishing a synthetic communication pathway initiated by self-destruction. Looking forward, this concept lays the foundation for the development of self-regulating artificial tissues, signal-propagating prototissues, and responsive material systems with controllable lifetimes and programmed disassembly.Author contributions
Conceptualization, J. K., and A. W.; methodology, J. K. and L. B. P.; writing – original draft, J. K.; writing – review and editing, J. K., L. B. P., and A. W.; funding acquisition, A. W.; resources, A. W.; supervision, A. W.Conflicts of interest
The authors declare no competing interests.Data availability
The datasets in the current study are available from the corresponding author on reasonable request. Supplementary information (SI): all experimental details, procedures, and supporting data for this article. See DOI: https://doi.org/10.1039/d5sc08882h.Acknowledgements
We acknowledge the financial support of the Deutsche Forschungsgemeinschaft (DFG) through the SFB 1066 project B14. L. B. P. acknowledges funding from the Alexander von Humboldt Foundation.References
- W. Park, S. Wei, B.-S. Kim, B. Kim, S.-J. Bae, Y. C. Chae, D. Ryu and K.-T. Ha, Exp. Mol. Med., 2023, 55, 1573–1594 CrossRef CAS PubMed.
- Y. Chen, X. Li, M. Yang and S.-B. Liu, Cell Death Dis., 2024, 15, 327 CrossRef PubMed.
- J. Yuan and D. Ofengeim, Nat. Rev. Mol. Cell Biol., 2024, 25, 379–395 CrossRef CAS.
- Y. Fuchs and H. Steller, Cell, 2011, 147, 742–758 CrossRef CAS PubMed.
- G. Kroemer, L. Galluzzi, O. Kepp and L. Zitvogel, Annu. Rev. Immunol., 2013, 31, 51–72 CrossRef CAS.
- S. Bedoui, M. J. Herold and A. Strasser, Nat. Rev. Mol. Cell Biol., 2020, 21, 678–695 CrossRef CAS PubMed.
- S. Maity, J. Ottelé, G. M. Santiago, P. W. J. M. Frederix, P. Kroon, O. Markovitch, M. C. A. Stuart, S. J. Marrink, S. Otto and W. H. Roos, J. Am. Chem. Soc., 2020, 142, 13709–13717 CrossRef CAS.
- D. Zwicker, R. Seyboldt, C. A. Weber, A. A. Hyman and F. Jülicher, Nat. Phys., 2017, 13, 408–413 Search PubMed.
- L. Olivi, M. Berger, R. N. P. Creyghton, N. De Franceschi, C. Dekker, B. M. Mulder, N. J. Claassens, P. R. ten Wolde and J. van der Oost, Nat. Commun., 2021, 12, 4531 Search PubMed.
- S. Tror, S. Jeon, H. T. Nguyen, E. Huh and K. Shin, Small Methods, 2023, 7, 2300182 Search PubMed.
- A. Belluati, S. Jimaja, R. J. Chadwick, C. Glynn, M. Chami, D. Happel, C. Guo, H. Kolmar and N. Bruns, Nat. Chem., 2024, 16, 564–574 CrossRef CAS PubMed.
- G. Cheng and J. Pérez-Mercader, Chem. Mater., 2019, 31, 5691–5698 CrossRef CAS.
- G. Cheng, S.-J. Hao, Y. Wan, D.-Y. Shan, J. Yang and S.-Y. Zheng, Chem. Mater., 2017, 29, 2081–2089 CrossRef CAS.
- S. Pearce and J. Perez-Mercader, ACS Cent. Sci., 2021, 7, 1543–1550 CrossRef CAS PubMed.
- H. Niederholtmeyer, C. Chaggan and N. K. Devaraj, Nat. Commun., 2018, 9, 5027 CrossRef PubMed.
- T.-Y. D. Tang, D. Cecchi, G. Fracasso, D. Accardi, A. Coutable-Pennarun, S. S. Mansy, A. W. Perriman, J. L. R. Anderson and S. Mann, ACS Synth. Biol., 2018, 7, 339–346 CrossRef CAS PubMed.
- A. O. Robinson, O. M. Venero and K. P. Adamala, Curr. Opin. Chem. Biol., 2021, 64, 165–173 CrossRef CAS PubMed.
- A. Joesaar, S. Yang, B. Bögels, A. Van Der Linden, P. Pieters, B. V. V. S. P. Kumar, N. Dalchau, A. Phillips, S. Mann and T. F. A. De Greef, Nat. Nanotechnol., 2019, 14, 369–378 CrossRef CAS PubMed.
- C. Qi, X. Ma, J. Zhong, J. Fang, Y. Huang, X. Deng, T. Kong and Z. Liu, ACS Nano, 2023, 17, 16787–16797 CrossRef CAS PubMed.
- A. Samanta, M. Hörner, W. Liu, W. Weber and A. Walther, Nat. Commun., 2022, 13, 3968 CrossRef CAS PubMed.
- J. Krehan, C.-R. Li, M. Masukawa, E. Amstad and A. Walther, Chem, 2025, 11(6), 102409 CAS.
- A. Diguet, M. Yanagisawa, Y.-J. Liu, E. Brun, S. Abadie, S. Rudiuk and D. Baigl, J. Am. Chem. Soc., 2012, 134, 4898–4904 CrossRef CAS PubMed.
- C. Sharma, I. Maity and A. Walther, Chem. Commun., 2023, 59, 1125–1144 RSC.
- T. Heuser, E. Weyandt and A. Walther, Angew. Chem., Int. Ed., 2015, 54, 13258–13262 CrossRef CAS PubMed.
- T. Heuser, R. Merindol, S. Loescher, A. Klaus and A. Walther, Adv. Mater., 2017, 29, 1606842 CrossRef PubMed.
- L. Heinen, T. Heuser, A. Steinschulte and A. Walther, Nano Lett., 2017, 17, 4989–4995 CrossRef CAS PubMed.
- X. Fan and A. Walther, Angew. Chem., Int. Ed., 2021, 60, 3619–3624 CrossRef CAS PubMed.
- X. Fan and A. Walther, Angew. Chem., Int. Ed., 2021, 60, 11398–11405 CrossRef CAS PubMed.
- I. Maity, C. Sharma, F. Lossada and A. Walther, Angew. Chem., Int. Ed., 2021, 60, 22537–22546 CrossRef CAS PubMed.
- Y. Miele, S. J. Jones, F. Rossi, P. A. Beales and A. F. Taylor, J. Phys. Chem. Lett., 2022, 13, 1979–1984 CrossRef CAS PubMed.
- Y. Miele, Z. Medveczky, G. Holló, B. Tegze, I. Derényi, Z. Hórvölgyi, E. Altamura, I. Lagzi and F. Rossi, Chem. Sci., 2020, 11, 3228–3235 RSC.
- D. Ridgway-Brown, A. S. Leathard, O. France, S. P. Muench, M. E. Webb, L. J. C. Jeuken, P. J. F. Henderson, A. F. Taylor and P. A. Beales, ACS Nano, 2025, 19, 9814–9825 CrossRef CAS PubMed.
- W. P. van den Akker, H. Wu, P. L. W. Welzen, H. Friedrich, L. K. E. A. Abdelmohsen, R. A. T. M. van Benthem, I. K. Voets and J. C. M. van Hest, J. Am. Chem. Soc., 2023, 145, 8600–8608 CrossRef CAS PubMed.
- H. Che, S. Cao and J. C. M. van Hest, J. Am. Chem. Soc., 2018, 140, 5356–5359 CrossRef CAS PubMed.
- X. Wang, S. Moreno, S. Boye, P. Wen, K. Zhang, P. Formanek, A. Lederer, B. Voit and D. Appelhans, Chem. Mater., 2021, 33, 6692–6700 CrossRef CAS.
- K. Zhang, S. Moreno, X. Wang, Y. Zhou, S. Boye, D. Voigt, B. Voit and D. Appelhans, Biomacromolecules, 2023, 24, 2489–2500 CrossRef CAS PubMed.
- P. Wen, X. Wang, S. Moreno, S. Boye, D. Voigt, B. Voit, X. Huang and D. Appelhans, Small, 2021, 17, 2005749 CrossRef CAS PubMed.
- D. Wang, S. Moreno, S. Boye, B. Voit and D. Appelhans, Chem. Commun., 2021, 57, 8019–8022 RSC.
- A. Sarkar, P. J. M. Swinkels, L. Duttenhofer, P. Besenius and A. Walther, Angew. Chem., Int. Ed., 2025, 64, e202503822 CrossRef CAS PubMed.
- J. Deng, D. Bezold, H. J. Jessen and A. Walther, Angew. Chem., Int. Ed., 2020, 59, 12084–12092 CrossRef CAS.
- J. S. Valera, Á. López-Acosta and T. M. Hermans, Angew. Chem., Int. Ed., 2024, 63, e202406931 CrossRef CAS PubMed.
- C. Bier, D. Binder, D. Drobietz, A. Loeschcke, T. Drepper, K.-E. Jaeger and J. Pietruszka, Synthesis, 2016, 49, 42–52 CrossRef.
- D. Binder, C. Bier, A. Grünberger, D. Drobietz, J. Hage-Hülsmann, G. Wandrey, J. Büchs, D. Kohlheyer, A. Loeschcke, W. Wiechert, K. Jaeger, J. Pietruszka and T. Drepper, ChemBioChem, 2016, 17, 296–299 CrossRef CAS PubMed.
- L. Gleue, B. Graefen, M. Voigt, J. Schupp, D. Schneider, M. Fichter, M. Kuske, V. Mailaender, A. Tuettenberg and M. Helm, ChemMedChem, 2025, 20, e202400648 CrossRef CAS PubMed.
- Y. Shimane and Y. Kuruma, Front. Bioeng. Biotechnol., 2022, 10, 873854 CrossRef PubMed.
- L. Baranda Pellejero, M. Vonk-de Roy, D. Baykal, L. Lehmann and A. Walther, J. Am. Chem. Soc., 2025, 147, 33643–33654 CrossRef CAS PubMed.
- S. Lotfipour Nasudivar, L. Pedrera and A. J. García-Sáez, Langmuir, 2024, 40, 25061–25068 CrossRef CAS PubMed.
- Y. Zheng, J. Sun, Z. Luo, Y. Li and Y. Huang, Cell Death Dis., 2024, 15, 859 CrossRef PubMed.
Footnote |
| † Lead contact. |
| This journal is © The Royal Society of Chemistry 2026 |