Open Access Article
Open Access ArticleInternally quenched fluorescent peptides provide insights into underexplored and reversible post-translational modifications
Jordi C. J.
Hintzen
a,
Kamiel D.
Beckley
a,
Emily L.
Goldberg
b and
George M.
Burslem
 *ac
*ac
aDepartment of Biochemistry and Biophysics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA. E-mail: George.Burslem@pennmedicine.upenn.edu
bDepartment of Physiology, University of California San Francisco, San Francisco, CA 94158, USA
cDepartment of Cancer Biology and Epigenetics Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA
First published on 5th December 2025
Abstract
Internally quenched fluorescent (IQF) peptides offer a powerful, modular platform for studying the enzymatic dynamics of post-translational modifications (PTMs) on lysine and arginine. Here we report a versatile IQF system that enables monitoring of PTM installation and removal via proteolytic cleavage by trypsin. This platform is compatible with both native PTMs and PTM mimetics, including acetylation, various other acylations, mono-/di-/trimethylation and citrullination across both histone and non-histone derived peptide substrates. Using synthetically accessible thialysine and thiaarginine analogs, we developed cysteine conjugation chemistries to access a wide array of PTM mimics, including novel reagents for lysine lactylation, β-hydroxybutyrylation and methyl-acetylation. Application of the system revealed distinct substrate preferences and site-specific activities for enzymes such as SIRT3, HDAC2, HDAC6, KDM3A, KDM4A and PAD4. Notably, the system uncovered enzymatic selectivity for acyl chain type and methylation state and demonstrated resistance of the emerging PTM methyl-acetyllysine to erasers. The system was also used to study the recently reported reversibility of acylation modifications by HDAC2 and 6 and is capable of evaluating enzymatic crosstalk between neighboring post-translational modifications. Our platform's adaptability and readout simplicity offer a generalizable chemical biology toolkit for PTM profiling, enzyme characterization, and inhibitor discovery.
Introduction
Post-translational modifications (PTMs) introduce an extraordinary level of complexity to the eukaryotic genome, playing essential roles in regulating protein function, localization, and stability.1 These modifications act as dynamic molecular switches that fine-tune a broad spectrum of cellular processes, ranging from gene expression and signal transduction to protein degradation and immune responses.2–7 Often these PTMs are small chemical changes installed on the amino acid side-chain, such as methylation, acetylation, and phosphorylation, but can expand to entire proteins being added to a residue such as in the case of ubiquitination.8 The dysregulation of PTMs has been implicated in numerous diseases, including neurodegenerative disorders such as Alzheimer's disease, cancer, and metabolic syndromes.9–12 PTMs on histone proteins, the proteins that constitute the nucleosome as the most basic unit of DNA compaction, have attracted significant attention due to their pivotal role in chromatin remodeling and regulation of gene expression, collectively giving rise to the field of epigenetics.3,13 In the epigenetic context, where different PTMs often occur on the same sites of histone tails, and a large amount of crosstalk exists between PTMs, the post-translational language becomes extremely complex.14 Despite the critical importance of these modifications, developing tools to study PTMs in a site-specific and quantitative manner remains a significant challenge.A major limitation in PTM research is the lack of specific tools, beyond mass spectrometry, to study the installation and removal of individual PTMs in isolation and in context of nearby modifications. Many techniques and assays lack the specificity to discriminate between structurally similar modifications, particularly when multiple PTMs can occur on the same amino acid residue. This complicates the analysis of enzymatic selectivity and kinetics for both the enzymes that install modifications, i.e., writers, and enzymes that remove these modifications, i.e., erasers. Inspired by the previous use of internally quenched fluorescence (IQF) peptides to study proteases15–21 and fluorophore peptide/acyl quencher pairs,22,23 we envisioned using IQF peptides as a versatile platform for studying PTMs in a site- and modification-specific manner. The IQF system is generated by preparing peptides bearing a quenching dinitrophenyl (Dnp) moiety in close proximity to a fluorescent aminobenzoyl (Abz) moiety, thus providing a “dark” peptide. Upon cleavage of the peptide by a protease, the Dnp is spatially removed from the aminobenzoyl moiety, turning on fluorescence (Fig. 1A).16,24 A commonly used peptidase to digest proteins is trypsin, a serine protease with specificity for cleaving after arginine and lysine residues.25 Conveniently, these are also two of the most commonly post-translationally modified amino acids, with lysine especially accommodating a wide array of PTMs.26–28 When a PTM is installed or removed from the cleavage site within the IQF peptide, enzymatic processing is expected to be altered, enabling direct assessment of the ability of writer and eraser enzymes to install or remove a modification at this site.
By using a two-step assay, both writers and erasers can be efficiently studied (Fig. 1A). In the case of writers, the unmodified substrate peptide is first incubated with the writer of interest. After a set reaction time, trypsin can be introduced into the mixture. We anticipated that only the portion of the peptide that remained unmodified will be cleaved, providing quantitative insight into the PTM installation. Conversely, when a post-translationally modified synthetic peptide is subjected to an eraser enzyme, the amount of cleavage by trypsin, or any other suitable peptidase, is a direct measure for the amount of PTM that the eraser enzyme has removed. These assays can therefore provide valuable insights into substrate preferences and catalytic efficiencies.
A challenge when employing this system is the synthetic accessibility of post-translationally modified amino acids that are directly amenable to solid-phase peptide synthesis (SPPS). While the most common modifications, such as lysine methylation, acetylation, and arginine citrullination, are commercially available, albeit at a much higher cost than unmodified lysine or arginine, more uncommon modifications need to be made synthetically and are therefore significantly less accessible for the generation of IQF peptides. To address this limitation, cysteine has been extensively studied as a chemical handle to install PTM mimics (Fig. 1B).29 Cysteine's thiol functionality provides a unique avenue for chemical derivatization using site-selective chemistry to generate a wide array of PTM analogs, including lysine acetylation and methylation. These bioconjugation reactions most often use simple and easy-to-use reagents to install these modifications. This allows for convenient and rapid access to various post-translationally modified synthetic peptides (Fig. 1B). In the context of this study, we aim to demonstrate the utility of this system through enzymatic assays with a variety of key writers and erasers involved in several different PTMs.
Results and discussion
Trypsin cleavage assays
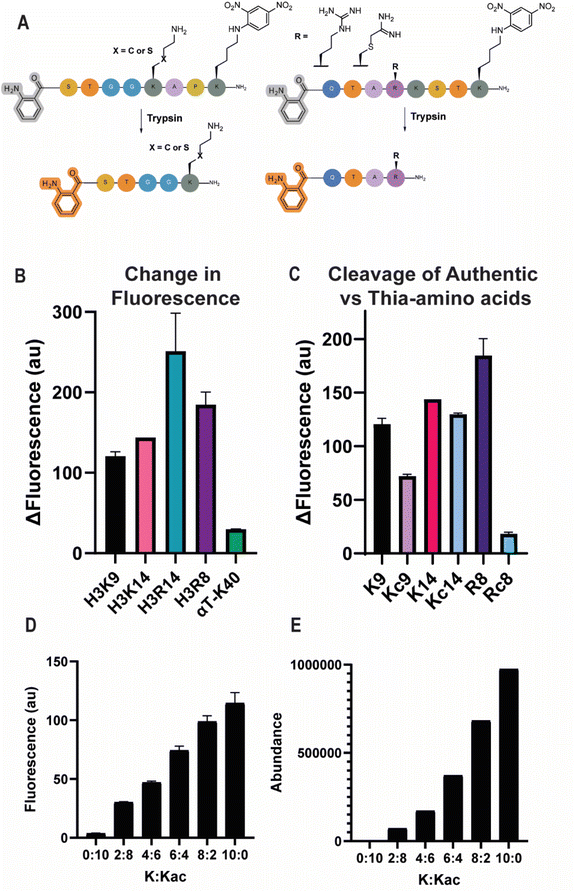
To establish the proof-of-concept for our fluorescent turn-on system, we performed trypsin cleavage assays on a series of histone H3-containing lysine and arginine residues (Fig. 2A). The panel of peptides was conveniently synthesized using standard Fmoc-based SPPS (Tables 1 and S1). Known as two major sites for post-translational modification of histone H3, we chose peptides based on the H3K9 and H3K14 sites,3 spanning residues 5–12 and 11–16 of histone H3, respectively, and these sequences were used for all peptides used to study lysine PTMs. In the case of arginine PTMs, site R8 was chosen as a known site for PAD4-mediated citrullination.30 Comparing the cleavage of H3K9, H3K14, and H3R8-containing peptides revealed notable differences in the amount and rate of trypsin cleavage, differing significantly depending on the sequence surrounding the basic residue (Fig. 2B and S1C). To analyze this further, lysine at position 14 was replaced by arginine to provide a more direct comparison between the two residues and here too, the amount of fluorescence signal and rate of cleavage were different between lysine and arginine, suggesting that both the nature of the cleaved amide and its surrounding environment play a major role in the efficiency of trypsin (Fig. 2B and S1C). To compare the trypsin cleavage of cysteine-derived mimics of lysine and arginine, thialysine was installed using 2-bromo-ethylamine hydrobromide and a thiaarginine mimic was installed by reacting cysteine with 2-chloroacetimidamide (CAM), which was synthesized from 2-chloroethanimidoate hydrochloride in one step (Fig. 3B).31–33 Importantly, thialysine and thiaarginine mimics were accepted efficiently as substrates in the assay. Cleavage of the thialysine mimics was found to be very comparable to the native lysine counterparts, while the thiaarginine peptide installed at position R8 showed a reduced amount of trypsin cleavage, it was still measurable and therefore viable for further enzymatic studies with PAD4 (Fig. 2C).31 To directly compare whether the observed fluorescence signal correlates well with the amount of lysine peptide present in a reaction mixture, different ratios of lysine and acetyllysine containing peptides were subjected to trypsin cleavage and analyzed both on the plate reader and a mass spectrometer, where the appearance of the Dnp-containing fragment could be efficiently tracked (Fig. S1). Importantly, the signals on both these instruments show a proportional increase that can be plotted and fitted using simple linear regression, thus validating our use of the plate reader-based assay to evaluate enzyme-catalyzed reactions with trypsin cleavage (Fig. 2D and E).| Enzyme | Peptides | Abbreviation |
|---|---|---|
| Trypsin | Abz-H3(6–12)K(Dnp) | H3K9 |
| Abz-H3(10–16)K(Dnp) | H3K14 | |
| Abz-H3(5–11)K(Dnp) | H3R8 | |
| Abz-H3(10–16)K(Dnp)K14R | H3R14 | |
| Abz-H3(6–12)K(Dnp)-Kc9 | H3Kc9 | |
| Abz-H3(10–16)K(Dnp)-Kc14 | H3Kc14 | |
| Abz-H3(5–11)K(Dnp)-Rc8 | H3Rc8 | |
| Abz-αT(37–43)K(Dnp) | αTK40 | |
| SIRT3/HDAC2 | Abz-H3(6–12)K(Dnp)-K9ac | H3K9ac |
| Abz-H3(6–12)K(Dnp)-K9lac | H3K9lac | |
| Abz-H3(6–12)K(Dnp)-K9bhb | H3K9bhb | |
| Abz-H3(6–12)K(Dnp)-Kc9ac | H3Kc9ac | |
| Abz-H3(6–12)K(Dnp)-Kc9lac | H3Kc9lac | |
| Abz-H3(6–12)K(Dnp)-Kc9bhb | H3Kc9bhb | |
| Abz-H3(10–16)K(Dnp)-K14ac | H3K14ac | |
| Abz-H3(10–16)K(Dnp)-K14lac | H3K14lac | |
| Abz-H3(10–16)K(Dnp)-K14bhb | H3K14bhb | |
| Abz-H3(10–16)K(Dnp)-Kc14ac | H3Kc14ac | |
| Abz-H3(10–16)K(Dnp)-Kc14lac | H3Kc14lac | |
| Abz-H3(10–16)K(Dnp)-Kc14bhb | H3Kc14bhb | |
| HDAC6 | Abz-αT(37–43)K(Dnp)-K40ac | αTK40ac |
| Abz-αT(37–43)K(Dnp)-Kc40ac | αTKc40ac | |
| KMD3A/4A | Abz-H3(6–12)K(Dnp)-K9me1 | H3K9me1 |
| Abz-H3(6–12)K(Dnp)-K9me2 | H3K9me2 | |
| Abz-H3(6–12)K(Dnp)-K9me3 | H3K9me3 | |
| Abz-H3(6–12)K(Dnp)-Kc9me1 | H3Kc9me1 | |
| Abz-H3(6–12)K(Dnp)-Kc9me2 | H3Kc9me2 | |
| Abz-H3(6–12)K(Dnp)-K9me3 | H3Kc9me3 | |
| Kmeac | Abz-H3(6–12)K(Dnp)-K9meac | H3K9meac |
| Abz-H3(6–12)K(Dnp)-Kc9meac | H3Kc9meac | |
| Abz-H3(10–16)K(Dnp)-K14meac | H3K14meac | |
| Abz-H3(10–16)K(Dnp)-Kc14meac | H3K14meac | |
| PAD4 | Abz-H3(5–11)K(Dnp) | H3R8 |
| Abz-H3(5–11)K(Dnp)-Rc8 | H3Rc8 | |
| Crosstalk | Abz-H3(6–12)K(Dnp)-S10ph | H3S10ph |
| Abz-H3(6–12)K(Dnp)-K9me2S10ph | H3K9me2S10ph | |
| Abz-H3(6–12)K(Dnp)-K9me3S10ph | H3K9me3S10ph | |
| Abz-H3(6–12)K(Dnp)-K9acS10ph | H3K9acS10ph |
Analog synthesis
To enable the study of post-translationally modified IQF peptides, we started with the synthesis of the Fmoc-Lys(lac)-OH building block, which was recently reported and could be repeated efficiently (Fig. S2).34 Encouraged by these results, Fmoc-Lys(bhb)-OH was synthesized in an analogous manner. Starting from the unprotected β-hydroxybutyric acid, the acid was first ethyl protected using sulfuric acid in ethanol and then followed the same transformations as for the lactyllysine analog to yield Fmoc-Lys(bhb)-OH in good yields overall. We then used these building blocks to synthesize the desired lactylated and β-hydroxybutyrylated peptides, while acetyllysine peptides were synthesized using commercially available building blocks. Starting from the tert-butyl protected intermediates from these syntheses, the vinyl amide reagents required for cysteine conjugation reactions were conveniently synthesized (Fig. S2). As was reported for a succinyllysine analog,35 the free acids could be converted to their respective amides using ethyl chloroformate and ammonium chloride in excellent yields. From here, the vinyl group was installed using vinyl iodide to give the final vinyl amide products, which could be directly used to conjugate to cysteine and after TFA deprotection of the tert-butyl protection groups yielded the thialysine-derived β-hydroxybutyrllysine and lactyllysine analogs Kcbhb and Kclac, respectively, directly on the peptide (Fig. S2).With the acetylated, β-hydroxybutyrylated and lactylated lysine and thialysine-derived peptides in hand, we continued with the synthesis of the rest of the panel of desired substrate peptides using previously reported conditions to conjugate to cysteine residues in peptides, while peptides containing native methylated and methyl-acetylated lysine residues were synthesized straightforwardly (Fig. 1B, S2 and Table 1).31,32,36 Methylated thialysine variants were generated by cysteine alkylation using halogenated ethylamine reagents, while acetylthialysine was installed using N-vinylacetamide in a radical initiated reaction akin to the chemistry employed for Kcbhb and Kclac.29,36 Conveniently, N-(2-chloroethyl)-N-methylacetamide could be synthesized by acetylation of 2-N-methylaminoethyl chloride hydrochloride and directly installed via cysteine alkylation to generate the desired Kcmeac peptides (Fig. 1B).
Evaluation of lysine acylation by IQF peptides
We started with the deacylase SIRT3 to study its ability to remove lysine acetylation (Kac), lysine lactylation (Klac), and lysine β-hydroxybutyrylation (Kbhb) based on SIRT3 being recently reported as a novel deacylase for the understudied Klac and Kbhb modifications at histone sites.37–40 SIRT3 was expressed recombinantly (Fig. S3), and its deacetylase activity was experimentally validated by using a p53(378–386)K382ac substrate peptide, which was readily deacetylated as observed by LC/MS after 1 hour under standard conditions (Fig. S4).41 To assess the ability of SIRT3 to remove acyl modifications from histone peptides, its activity was tested on peptides modified with Kac, Klac, and Kbhb at the H3K9 and H3K14 sites (Fig. 3A). SIRT3 efficiently removed Kac and Klac from the K9 position, but interestingly, it was only able to remove Kac from the K14 site, contrary to reported results (Fig. 3C and D).40 This could be due to limitations of our experimental system, if, for instance, efficient removal of lactylation from the H3K14 site by SIRT3 requires a larger portion of its histone substrate to be present, or if additional factors are needed to ensure efficient delactylation at this site. Moreover, in this assay, SIRT3 was not able to remove Kbhb from either the K9 or K14 site, suggesting that the Kbhb substrate is less tolerated by the enzyme compared to Kac and Klac (Fig. 3C and D).39 We observed similar results with the thialysine-derived peptides, further adding to the possibility that H3K9 is the preferred deacylation site for SIRT3 (Fig. 3C and D). In these experiments, Kcac and Kclac were removed from the H3K9 site, while at the H3K14 position, even acetylated thialysine was not a substrate of SIRT3. Although, it was previously reported that acetylated thialysine is removed by SIRT2 at the H4K16 site suggesting this is not solely a feature of the thialysine moiety.36 Lastly, the thialysine-derived mimic of Kbhb, Kcbhb, was not cleaved from either the H3K9 or H3K14 sites (Fig. 3C and D). These findings highlight subtle differences in SIRT3 substrate preference, both in terms of site specificity and modification type, and showcases the potential of our system to probe enzyme selectivity site specifically.Next, we performed similar analyses with HDAC2-catalyzed deacylation as a model enzyme for the Zn2+-dependent histone deacetylase family (Fig. 3A).42 Delactylation activity was recently reported for class I HDACs,43 and our laboratories recently showed that HDAC2 can reversibly catalyze lactylation and β-hydroxybutyrylation on histone sites under high, but physiological, lactate or β-hydroxybutyrate concentrations.44,45 We used our system to analyze this catalytic behavior in both the writer and eraser direction for HDAC2 in detail (Fig. 3A and B). Analogous to the assays with SIRT3, acyl-modified peptides carrying an acetyl, lactyl and β-hydroxybutyryl modification at positions K9 and K14 were incubated with recombinantly expressed HDAC2 and subsequently their cleavage by trypsin was measured (Fig. 3E and F). Interestingly, like SIRT3, HDAC2 was able to efficiently remove the acetyl and lactyl marks from the K9 position, as is evident from the increased fluorescence in these samples but appeared completely inert towards the K14 site in these assays as well as β-hydroxybutyrylation at both positions.
Next, the ability of HDAC2 to install acyl modifications was investigated in this system, following recent reports from our groups and others.44–46 After initial incubation with 5 mM acetate, L-lactate, or β-hydroxybutyrylate with HDAC2 (Fig. 3G), lactylation appeared to be by far the most efficiently installed modification. It is important to note that unlike eraser studies, the ability of a writer enzyme to install a modification is indicated by a complete loss of fluorescence signal and the subsequent inability of trypsin to cleave at this site. This is seen at both the K9 and K14 sites when HDAC2 is incubated with lactate. Encouraged by the lactylation results, we tested increasing concentrations of acetate and β-hydroxybutyrylate with HDAC2 (Fig. 3H). Within this system, no significant acetylation was observed, while on the other hand, a reduction in fluorescence cleavage was observed when increasing the β-hydroxybutyryate concentration to 100 mM, indicating that β-hydroxybutyrylation can be catalyzed by HDAC2 in this system, albeit which much lower efficiency compared to lactylation, verifying the preferences observed in other biochemical and cellular assays.44,45 We then expanded our evaluation of the HDAC2-catalyzed acylation modifications by incubating HDAC2 with a variety of short-chain fatty acids at 10 mM (Fig. S5) and 100 mM (Fig. 3H). Notably, the only modification to show significant activity at 10 mM in these assays is lactylation, while, excitingly, we show that HDAC2 can catalyze in vitro generation of propionylation, crotonylation, 2-hydroxyisobutyrylation and lipoylation at 100 mM of these compounds present (Fig. 3I). While HDAC2-catalyzed propionylation and crotonylation have already been reported, 2-hydroxyisobutyrylation and lipoylation are exciting new modifications that could potentially be installed by class I HDACs under physiologically relevant conditions.45,47 We next applied the system to investigate its potential to discover and benchmark inhibitors for studied enzymes. In this case, we used the well-known deacetylase inhibitor SAHA (Voronistat) and incubated it at different concentrations in the HDAC2-catalyzed K9 deacetylation reactions (Fig. S6). Gratifyingly, an IC50 of 59.23 nM was observed, which is well in line with reported IC50 values for this compound.48
Finally, to extend our study of deacetylases to substrates beyond histone proteins, HDAC6, a member of the Class IIb HDACs was identified as an interesting target given it's localization to the cytosol.49,50 HDAC6 is an important microtubule-associated deacetylase, removing the acetyl mark from α-tubulin K40 (αT-K40), where it plays roles in microtubule associated cellular movement.49,51 Interestingly, HDAC6, like HDAC2, was reported to have reversible activity, where it is able to lactylate the αT-K40 site.46 To study the installation and removal of acyl modifications at the αT-K40 site by HDAC6, we synthesized a set of peptides carrying unmodified lysine, acetyllysine and thiacetyllysine. Like the H3-derived peptides, αT-K40 showed a measurable fluorescence increase when incubated with trypsin (Fig. 2B). The catalytic domain of HDAC6 was expressed and purified (Fig. S7)52 and we then carried out trypsin coupled assays analysing the deacetylation of an acetylated lysine and thialysine αT-K40 peptide as well as lactylation of αT-K40 by HDAC6 (Fig. S8). In line with our results for SIRT3, HDAC6 was able efficiently deacetylate the αT-K40ac peptide, while the thialysine variant, αT-Kc40ac was inert to the presence of the enzyme (Fig. S8a). Furthermore, we were able to show that in our assay context, HDAC6 is able to fully catalyze the lactylation of αT-K40 at 100 mM of L-lactate present, while at 10 mM moderate lactylation could be observed, corroborating that the lactylation of HDAC6 can occur at physiologically relevant concentrations of lactate (Fig. S8b).46
Evaluation of lysine demethylation and arginine citrullination by IQF peptides
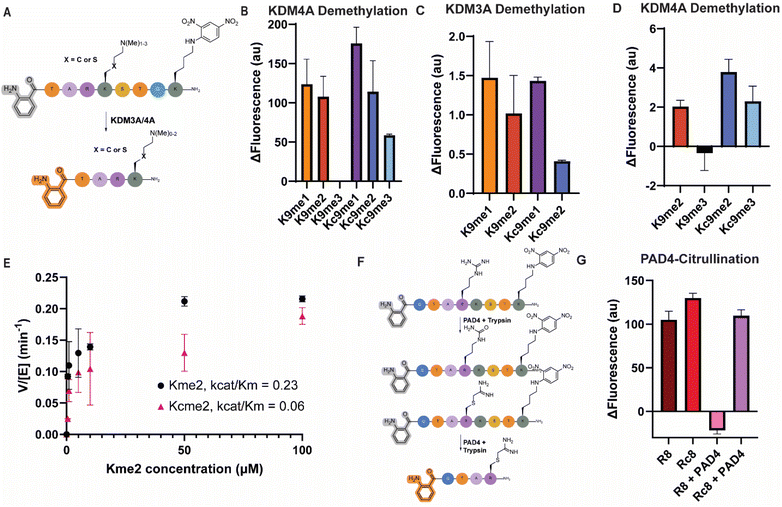
Beyond acylation, we used the lysine demethylases KDM3A and KDM4A to investigate the ability of trypsin to cleave after methylated lysine residues, both in their native and thia-lysine derived state. We explored the influence of lysine methylation state on trypsin cleavage by analyzing histone H3K9 peptides with mono-, di-, and trimethyl modifications (Fig. 4A). As expected, higher methylation states conferred increased resistance to cleavage, with the trimethyllysine-modified peptide exhibiting near-complete stability over the course of this experiment (Fig. 4B).53 Notably, this trend was consistent across both native methyllysine residues and thialysine-derived mimics (Fig. 4B), confirming the applicability of methylated thialysines as trypsin substrates, making them amenable to our system for studying methylation-dependent proteolysis by KDMs. Surprisingly, a small amount of cleavage was observed when the thialysine-derived Kcme3 modification was introduced (Fig. 4B). However, when looking at the progress of the reaction in real-time, it can clearly be observed that the rate of cleavage for both the methylated lysine and thialysine peptides convincingly follows a trend where rate of cleavage is reduced with increased methylation state (Fig. S9A and B).To further investigate methylation dynamics, demethylation assays were performed with KDM3A and KDM4A. KDM3A is reported to demethylate mono- and dimethyllysine at the H3K9 position, while KDM4A catalyzes the demethylation of di- and trimethylation at the same site.54 We were able to detect demethylation by both KDM3A and KDM4A in this assay (Fig. 4C and D), as cleavage rates increased upon incubation with the respective KDMs. Strikingly, the trimethylated reaction remained completely inert to trypsin cleavage after incubation with KDM4A, while for the dimethylated peptide, cleavage was readily observed (Fig. 4D). These subtle changes suggest that the demethylation reactions were not complete under our assay conditions, leading to a mixture of methylation states present in the samples at the point of addition of trypsin. Notably, the behavior of thialysine-derived methyl mimics differed slightly (Fig. 4C and D). KDM3A showed a reduced demethylation activity on the Kcme2 peptide, while KDM4A seemingly shows a preference for the thialysine-derived peptides, with clear fluorescence cleavage observed when incubated with Kcme3 and an increased fluorescence cleavage with Kcme2 (Fig. 4C and D). Previously it was reported that KDM4A could indeed demethylate thialysine at H3K9 all the way down to the unmethylated state, albeit in the context of full nucleosomes, suggesting that like SIRT3, additional substrate recognition might be needed for full enzymatic activity.55 To compare the demethylation of thialysine and lysine peptides by these demethylases in more detail, we measured Michaelis–Menten kinetics on the KDM3A-catalyzed demethylation of dimethylated substrates. Due to the sequential reactions KDM3A can carry out, we carefully ensured that only the reaction from the dimethyl to the monomethyl state was being evaluated, by directly comparing to cleavage rates of the authentic monomethyl peptides themselves, giving us a handle to calculate the conversions (Fig. 4E). These kinetics assays resulted in an observed kcat of 0.19 ± 0.02 and 0.16 ± 0.03 min−1 for the Kme2 and Kcme2 peptides, respectively, while the Kms were found to be 0.81 ± 0.29 and 2.53 ± 1.61 for Kme2 and Kcme2, respectively. Our calculated kcat/Km values showed an apparent four times faster demethylation rate for Kme2 compared to Kcme2, mainly driven by a reduced Km, which indicates a less preferred mode of binding to the enzyme active site. Our results agree with prior studies that evaluated Michaelis–Menten kinetics of thialysine-derived histone peptides when studying lysine methylases, acetylases and deactelyases.56,57
PAD4 coupled cleavage assays
Next, the impact of PAD4-mediated citrullination of H3R8 was evaluated on trypsin cleavage (Fig. 4F). Citrullination of the arginine residue effectively blocked cleavage, demonstrating the system's ability to detect citrullination events (Fig. 4G). However, when we installed the thiaarginine mimic at this site and incubated with PAD4, trypsin could still effectively cleave the H3Rc8 peptide, indicating that PAD4 does not accept this modification as a substrate (Fig. 4G). Likely, the structure is too chemically different from arginine, since it has one atom less in its sidechain, lacks a guanidinium nitrogen, and contains a C–S–C instead of a C–C–C backbone. This provides an interesting opportunity for installation of this arginine mimic in cases where blocking of protein citrullination could have a therapeutic effect, for example in the case of amyloid-β peptides in Alzheimer's disease.58Investigation of removal of Kmeac by IQF peptides
It was recently discovered that lysine can be methylated and acetylated at the same site simultaneously (Kmeac),59 while epigenetic proteins that write, erase or read this modification are still unknown. Combining our HDAC and KDM assays, we investigated whether the Kmeac PTM can be effectively removed by either class of enzymes in our model system. Additionally, convenient cysteine conjugation chemistry was developed to install the Kmeac modification via thialysine (Fig. 1B). Using our IQF system and the enzymes studied so far, we sought to identify whether Kmeac demethylation or deacetylation can be catalyzed by KDM3A, KDM4A, SIRT3 and HDAC2, respectively. Initially, no appreciable rise in fluorescence was observed for any of the Kmeac or Kcmeac peptides after incubation with the enzymes (Fig. S10A and B). However, even after potential demethylation by KDM3A and KDM4A, no fluorescence increase would be observed since acetylated lysine is not cleaved by trypsin. Thus, enzymatic reactions were subjected to LC-MS analysis, which revealed that indeed, the Kmeac modification appears inert to this set of enzymes within this experimental system, verifying that our approach could be used as a first pass screen to identify editors of newly emerging PTMs (Fig. S11A and B).Studying enzymatic crosstalk using IQF peptides
Histone tails are well known to get decorated with a complex array of PTMs. Among these, many are installed on neighboring residues, and mounting evidence suggests that significant crosstalk exists between serine phosphorylation on one residue and lysine methylation or acetylation on a nearby residue.60–63 We applied our IQF system to assess whether phosphorylation at the H3S10 site affects removal of methylation and acetylation marks on the H3K9 site and synthesized four peptides containing a phosphorylation at the serine 10 and lysine either unmodified, dimethylated, trimethylated or acetylated. Initially, we compared the trypsin cleavage of the H3K9 peptide to the H3K9S10ph peptide and saw that the rate of cleavage is only marginally affected by introducing phosphorylation (Fig. 5A). We then proceeded to incubate the lysine modified peptides with their respective eraser enzymes. For the H3K9acS10ph we observed that both in the case of SIRT3 and HDAC2 incubation, the amount of trypsin cleavage was higher, indicating that HDAC activity is accelerated when S10ph is present, while for the H3K9me2S10ph, incubated with KDM3A, and the H3K9me3S10ph, incubated with KDM4A, the amount of trypsin cleavage was lower compared to the non-phosphorylated counterpart (Fig. 5B). These findings suggest there might be significant post-translational crosstalk between the S10 and K9 sites, with phosphorylation either increasing or decreasing the rates of removal of different PTMs at K9 and verify the use of IQF peptides to study PTM crosstalk.Conclusions
In conclusion, we have developed a robust and adaptable fluorescent turn-on system capable of detecting changes in PTMs through enzymatic cleavage assays. This system provides a powerful tool for monitoring these PTM states and has been shown to offer wide applicability across different enzymatic processes and modification types. Through these investigations, nuanced differences were revealed in substrate preferences of key enzymes, such as SIRT3, HDAC2, HDAC6, PAD4, KDM3A, and KDM4A, providing valuable insights into their specificities and catalytic behaviors. In addition, new bioconjugation chemistry was developed to install thialysine mimics of Kmeac, Klac and Kbhb, expanding the repertoire of accessible PTM analogs. A key strength of this approach lies in the ease of use of thialysine chemistry, which offers a straightforward, cost-effective, and efficient alternative to the synthesis or purchase of native PTM amino acids. This enables rapid and flexible generation of modified peptides without the need for extensive synthetic efforts. Additionally, cysteine-derived PTM mimics are easily accessible on the full protein level as well, which can be achieved by site-directed mutagenesis to install a cysteine residue, making PTM studies more accessible.Our results demonstrated that SIRT3 discriminates between acyl modifications at different histone sites, preferentially removing Kac and Klac over Kbhb and exhibiting site specificity. While for HDAC2, it was observed that acetylation and lactylation could be removed depending on the histone site and lactylation was efficiently catalyzed under high lactate concentrations. HDAC2 was also shown to catalyze the formation of 2-hydroxyisobutyrylated and lipoylated lysine on a histone substrate, which are novel findings in vitro that are currently subject to further validation. Finally, HDAC6 was shown to efficiently remove an acetyl and install a lactyl mark at the αT-K40 site using our IQF system.
Similarly, KDM3A and KDM4A displayed distinct demethylation patterns, which were further elucidated through the use of thialysine-derived PTM mimics. Provisionally, we have also shown that Kmeac is inert to demethylation and deacetylation within this simple experimental system (at least to the enzymes tested). We could observe that PAD4 can efficiently catalyze citrullination of arginine, but not of a thiaarginine mimic. The application of thialysine analogs proved particularly insightful, as they served as reliable substitutes for native PTMs while revealing subtle differences in enzyme activity that may not be apparent with canonical amino acids. Additionally, the system allowed us analyze PTM crosstalk on neighboring residues on histone tail peptides, potentially providing important insights into the interplay between PTMs in an epigenetic context.
Overall, this system not only facilitates the study of individual PTMs and their effects on enzymatic processing but also provides a modular platform for expanding into more complex PTM landscapes. By employing synthetic peptide substrates with site-specific modifications, we can now systematically dissect the roles of PTMs in epigenetic regulation and beyond. Furthermore, the versatility of this system opens the door to extending the platform to other proteases, enabling the study of PTMs on a wider variety of amino acids beyond lysine and arginine. This work establishes a foundation for future studies aiming to unravel the intricate regulatory networks controlled by PTMs and to develop targeted inhibitors or probes for PTM-associated enzymes.
Materials and methods
Peptide synthesis and purification
Peptides were synthesized by manual Fmoc solid phase peptide chemistry on Rink-Amide MBHA resin. The resin underwent iterative cycles of Fmoc deprotection (20% piperidine in DMF, 30 minutes), amino acid coupling (4 eq. Fmoc AA, 4 eq. HATU in 4% DIPEA in DMF, 1 hour) and washing (3 × DMF) until completion of the sequences. The N-terminal Abz group was installed by coupling with 5 eq. Boc-2-Abz-OH, 5 eq. HATU and 10 eq. of DIPEA in DMF for 1 hour. Peptides were cleaved from the resin in a mixture of TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIPS
TIPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O at the ratio of 95
H2O at the ratio of 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 for 3 hours at room temperature, for peptides containing a cysteine residue a mixture of TFA
5 for 3 hours at room temperature, for peptides containing a cysteine residue a mixture of TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIPS
TIPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDT at the ratio of 92.5
EDT at the ratio of 92.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 was used. The peptides were precipitated using ice-cold diethyl ether, centrifuged at 4500 rpm at 4 °C for 5 minutes, redissolved in a mixture of MQ-H2O and MeCN and purified by semi-preparative reverse phase chromatography eluting with 5 to 95% MeCN in H2O containing over a C18 column. Peaks containing peptides were identified by LC-MS, pooled, and concentrated in vacuo to yield white or bright yellow solids. HRMS analyses were conducted on an Agilent 1290 Infinity II UHPLC system coupled with an Agilent 6545XT AdvanceBio LC/Q-TOF system equipped with an Agilent Dual Jet Stream ESI source.
2.5 was used. The peptides were precipitated using ice-cold diethyl ether, centrifuged at 4500 rpm at 4 °C for 5 minutes, redissolved in a mixture of MQ-H2O and MeCN and purified by semi-preparative reverse phase chromatography eluting with 5 to 95% MeCN in H2O containing over a C18 column. Peaks containing peptides were identified by LC-MS, pooled, and concentrated in vacuo to yield white or bright yellow solids. HRMS analyses were conducted on an Agilent 1290 Infinity II UHPLC system coupled with an Agilent 6545XT AdvanceBio LC/Q-TOF system equipped with an Agilent Dual Jet Stream ESI source.
Cysteine conjugation reactions
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 736; RRID:Addgene_13736). The SIRT3 construct was expressed according to previously published methods.41 Briefly, SIRT3 was expressed in BL21(DE3) E. coli cells in YT medium supplemented with 100 µg mL−1 ampicillin. Cells were grown at 37 °C in a shaker to an OD600 of 0.6–0.8 before inducing with 0.5 mM isopropyl βD-1-thiogalactopyranoside (IPTG) and expressing at 37 °C for 16 hours before collecting cells by centrifugation. Cell pellets were resuspended in 50 mM Tris buffer pH 8.0 containing 250 mM NaCl, 5 mM imidazole, 1 mM β-mercaptoethanol and protease inhibitor cocktail. Cells were lysed by sonication on ice and lysates were cleared by centrifugation at 13
736; RRID:Addgene_13736). The SIRT3 construct was expressed according to previously published methods.41 Briefly, SIRT3 was expressed in BL21(DE3) E. coli cells in YT medium supplemented with 100 µg mL−1 ampicillin. Cells were grown at 37 °C in a shaker to an OD600 of 0.6–0.8 before inducing with 0.5 mM isopropyl βD-1-thiogalactopyranoside (IPTG) and expressing at 37 °C for 16 hours before collecting cells by centrifugation. Cell pellets were resuspended in 50 mM Tris buffer pH 8.0 containing 250 mM NaCl, 5 mM imidazole, 1 mM β-mercaptoethanol and protease inhibitor cocktail. Cells were lysed by sonication on ice and lysates were cleared by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 45 minutes. SIRT3 was further purified by Ni-NTA column purification, dialyzed into 50 mM Tris buffer, pH 8.0 containing 150 mM NaCl and 1 mM MgCl2 and concentrated to a 318 µM concentration. The purified protein was aliquoted before before flash-freezing in liquid nitrogen and storing at −80 °C for future use.
000×g for 45 minutes. SIRT3 was further purified by Ni-NTA column purification, dialyzed into 50 mM Tris buffer, pH 8.0 containing 150 mM NaCl and 1 mM MgCl2 and concentrated to a 318 µM concentration. The purified protein was aliquoted before before flash-freezing in liquid nitrogen and storing at −80 °C for future use.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 45 minutes. HDAC6 was further purified by Ni-NTA column purification, dialyzed with 6 mg mL−1 TEV protease, further purified by size exclusion chromatography and finally concentrated to a 94 µM concentration. The purified protein was aliquoted before before flash-freezing in liquid nitrogen and storing at −80 °C for future use.
000×g for 45 minutes. HDAC6 was further purified by Ni-NTA column purification, dialyzed with 6 mg mL−1 TEV protease, further purified by size exclusion chromatography and finally concentrated to a 94 µM concentration. The purified protein was aliquoted before before flash-freezing in liquid nitrogen and storing at −80 °C for future use.
General methods for fluorescence turn-on experiments
The cleavage of the Abz/Dnp-substrates was monitored by measuring the fluorescence intensity in a Varioskan LUX microplate reader at emission and excitation wavelengths of 320 and 420 nm, respectively, each minute over 1 hour. Triplicate measurements were taken for each data point. The fluorescence data was processed using GraphPad Prism. The data were reported as mean ± SE%.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456 & 31
456 & 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 457). KDM-catalyzed reactions were performed in a 300 µL final volume in 50 mM HEPES buffer at pH 7.5 with 100 µM LAA, 10 µM FAS, 10 µM 2-OG. 10 µM of peptide were incubated with 1 µM KDM for 1 hour at 37 °C. Subsequently, reaction mixtures were transferred to 384 well Corning NBS microplates (flat bottom, no lid, low flange, non-binding surface, non-sterile, black) and trypsin was added to a final concentration of 1 µg mL−1. For the kinetics measurements varying concentrations of Kme2 or Kcme2 peptides were used, and the data were analyzed using nonlinear regression according to Michaelis–Menten kinetics.
457). KDM-catalyzed reactions were performed in a 300 µL final volume in 50 mM HEPES buffer at pH 7.5 with 100 µM LAA, 10 µM FAS, 10 µM 2-OG. 10 µM of peptide were incubated with 1 µM KDM for 1 hour at 37 °C. Subsequently, reaction mixtures were transferred to 384 well Corning NBS microplates (flat bottom, no lid, low flange, non-binding surface, non-sterile, black) and trypsin was added to a final concentration of 1 µg mL−1. For the kinetics measurements varying concentrations of Kme2 or Kcme2 peptides were used, and the data were analyzed using nonlinear regression according to Michaelis–Menten kinetics.
Synthetic methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). After stirring at room temperature for 16 hours, the solution was further dilute with water and washed with petroleum ether twice. Then, the mixture was acidified to pH 2 with 1 M HCl and extracted with DCM three times. The organic phase was dried over over MgSO4, filtered and concentrated in vacuo to afford (S)-2-(tert-butoxy)propanoic acid (390 mg, 2.7 mmol, 88%). 1H NMR (400 MHz, CDCl3): JH1-60 MS calcd for C7H13O3Na+ [M + Na]+, 169.1; found 169.0.
1, v/v). After stirring at room temperature for 16 hours, the solution was further dilute with water and washed with petroleum ether twice. Then, the mixture was acidified to pH 2 with 1 M HCl and extracted with DCM three times. The organic phase was dried over over MgSO4, filtered and concentrated in vacuo to afford (S)-2-(tert-butoxy)propanoic acid (390 mg, 2.7 mmol, 88%). 1H NMR (400 MHz, CDCl3): JH1-60 MS calcd for C7H13O3Na+ [M + Na]+, 169.1; found 169.0.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) to afford ethyl 3-(tert-butoxy)butanoate (450 mg, 2.4 mmol, 68%). 1H NMR (400 MHz, CDCl3) δ 4.05–3.86 (m, 2H), 2.32 (dd, J = 14.5, 6.3 Hz, 1H), 2.17 (dd, J = 14.5, 7.0 Hz, 1H), 1.09 (td, J = 7.0, 3.1 Hz, 3H), 1.06–0.96 (m, 12H). 13C NMR (101 MHz, CDCl3) δ 171.37, 73.44, 64.54, 59.90, 44.10, 28.14, 23.14, 14.01. MS calcd for C10H21O3+ [M + H]+, 189.14; found JH1-71.
60) to afford ethyl 3-(tert-butoxy)butanoate (450 mg, 2.4 mmol, 68%). 1H NMR (400 MHz, CDCl3) δ 4.05–3.86 (m, 2H), 2.32 (dd, J = 14.5, 6.3 Hz, 1H), 2.17 (dd, J = 14.5, 7.0 Hz, 1H), 1.09 (td, J = 7.0, 3.1 Hz, 3H), 1.06–0.96 (m, 12H). 13C NMR (101 MHz, CDCl3) δ 171.37, 73.44, 64.54, 59.90, 44.10, 28.14, 23.14, 14.01. MS calcd for C10H21O3+ [M + H]+, 189.14; found JH1-71.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). After stirring at room temperature for 16 hours, the solution was further dilute with water and washed with petroleum ether twice. Then, the mixture was acidified to pH 2 with 1 M HCl and extracted with DCM three times. The organic phase was dried over over MgSO4, filtered and concentrated in vacuo to afford 3-(tert-butoxy)butanoic acid (289 mg, 1.8 mmol, 75%). 1H NMR (400 MHz, CDCl3) δ 4.04 (h, J = 6.1 Hz, 1H), 2.47 (dd, J = 14.9, 5.8 Hz, 1H), 2.36 (dd, J = 14.9, 7.0 Hz, 1H), 1.16 (m, 12H). 13C NMR (101 MHz, CDCl3) δ 176.47, 74.30, 64.59, 43.76, 28.15, 23.06. MS calcd for C8H17O3 [M + H]+, 183.1; found 183.0.
1, v/v). After stirring at room temperature for 16 hours, the solution was further dilute with water and washed with petroleum ether twice. Then, the mixture was acidified to pH 2 with 1 M HCl and extracted with DCM three times. The organic phase was dried over over MgSO4, filtered and concentrated in vacuo to afford 3-(tert-butoxy)butanoic acid (289 mg, 1.8 mmol, 75%). 1H NMR (400 MHz, CDCl3) δ 4.04 (h, J = 6.1 Hz, 1H), 2.47 (dd, J = 14.9, 5.8 Hz, 1H), 2.36 (dd, J = 14.9, 7.0 Hz, 1H), 1.16 (m, 12H). 13C NMR (101 MHz, CDCl3) δ 176.47, 74.30, 64.59, 43.76, 28.15, 23.06. MS calcd for C8H17O3 [M + H]+, 183.1; found 183.0.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and 3-bromopropene (0.47 mL, 5.5 mmol, 1.1 eq.) and Na2CO3 (585 mg, 5.5 mmol) were added. The reaction was stirred at room temperature for 16 hours after which the reaction was diluted with water and extracted with EtOAc three times. The organic phase was washed with water five times and dried of MgSO4, filtered and concentrated in vacuo to afford Fmoc-Lys(Boc)-OAllyl (2.40 g, 4.73 mmol, 95%) which was used without further purification.
1, v/v) and 3-bromopropene (0.47 mL, 5.5 mmol, 1.1 eq.) and Na2CO3 (585 mg, 5.5 mmol) were added. The reaction was stirred at room temperature for 16 hours after which the reaction was diluted with water and extracted with EtOAc three times. The organic phase was washed with water five times and dried of MgSO4, filtered and concentrated in vacuo to afford Fmoc-Lys(Boc)-OAllyl (2.40 g, 4.73 mmol, 95%) which was used without further purification.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and stirred for 2 hours at room temperature and the reaction was directly concentrated in vacuo. The crude Fmoc-Lys(NH2)-OAllyl (1.93 g, 4.73 mmol, 100% yield) was used without further purification. MS calcd for C24H29N2O4 [M + H]+, 409.2; found 409.3.
1, v/v) and stirred for 2 hours at room temperature and the reaction was directly concentrated in vacuo. The crude Fmoc-Lys(NH2)-OAllyl (1.93 g, 4.73 mmol, 100% yield) was used without further purification. MS calcd for C24H29N2O4 [M + H]+, 409.2; found 409.3.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70) to afford Fmoc-Lys(lac)-OAllyl (570 mg, 1.0 mmol, 100%). MS calcd for C31H41N2O6 [M + H]+, 537.3; found 537.2.
70) to afford Fmoc-Lys(lac)-OAllyl (570 mg, 1.0 mmol, 100%). MS calcd for C31H41N2O6 [M + H]+, 537.3; found 537.2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford Fmoc-Lys(lac)-OH (540 mg, 1.1 mmol, 87%). 1H NMR (400 MHz, CDCl3) δ 7.74 (d, J = 7.5 Hz, 2H), 7.70–7.60 (m, 2H), 7.60–7.52 (m, 1H), 7.50–7.43 (m, 1H), 7.37 (t, J = 7.5 Hz, 2H), 7.28 (td, J = 7.4, 1.3 Hz, 2H), 4.48–4.30 (m, 1H), 4.19 (t, J = 7.1 Hz, 1H), 4.16–4.03 (m, 3H), 3.26 (p, J = 6.8 Hz, 1H), 2.04 (s, 3H), 1.31 (d, J = 6.9 Hz, 2H), 1.25 (t, J = 7.1 Hz, 3H), 1.18 (s, 7H). MS calcd for C28H36N2O6 [M + H]+, 497.3; found 497.3.
1) to afford Fmoc-Lys(lac)-OH (540 mg, 1.1 mmol, 87%). 1H NMR (400 MHz, CDCl3) δ 7.74 (d, J = 7.5 Hz, 2H), 7.70–7.60 (m, 2H), 7.60–7.52 (m, 1H), 7.50–7.43 (m, 1H), 7.37 (t, J = 7.5 Hz, 2H), 7.28 (td, J = 7.4, 1.3 Hz, 2H), 4.48–4.30 (m, 1H), 4.19 (t, J = 7.1 Hz, 1H), 4.16–4.03 (m, 3H), 3.26 (p, J = 6.8 Hz, 1H), 2.04 (s, 3H), 1.31 (d, J = 6.9 Hz, 2H), 1.25 (t, J = 7.1 Hz, 3H), 1.18 (s, 7H). MS calcd for C28H36N2O6 [M + H]+, 497.3; found 497.3.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70) to afford Fmoc-Lys(bhb)-OAllyl (466 mg, 0.8 mmol, 84%). 1H NMR (400 MHz, CDCl3) δ 7.66 (d, J = 7.5 Hz, 2H), 7.53 (t, J = 6.6 Hz, 2H), 7.33–7.26 (m, 2H), 7.20 (tt, J = 7.4, 0.9 Hz, 2H), 5.81 (ddt, J = 16.1, 11.0, 5.7 Hz, 1H), 5.23 (dd, J = 17.3, 1.6 Hz, 1H), 5.14 (dd, J = 10.4, 1.1 Hz, 1H), 4.54 (d, J = 5.8 Hz, 2H), 4.13 (t, J = 7.2 Hz, 1H), 4.01 (q, J = 7.1 Hz, 2H), 3.95 (q, J = 6.0 Hz, 1H), 3.25–3.03 (m, 1H), 2.27–2.12 (m, 2H), 1.88–1.60 (m, 1H), 1.53–1.34 (m, 2H), 1.12–1.05 (m, 12H), 0.84–0.70 (m, 4H). 13C NMR (101 MHz, CDCl3) δ 162.44, 141.17, 131.57, 127.60, 126.97, 119.85, 118.60, 73.89, 65.72, 64.80, 60.23, 53.84 (d, J = 5.1 Hz), 47.08, 45.71, 38.48, 36.34, 31.27, 28.96 (d, J = 6.9 Hz), 28.27, 22.82 (d, J = 1.8 Hz), 22.62, 22.54, 20.90, 14.06 (d, J = 7.3 Hz). MS calcd for C32H43N2O6 [M + H]+, 552.3; found 552.3.
70) to afford Fmoc-Lys(bhb)-OAllyl (466 mg, 0.8 mmol, 84%). 1H NMR (400 MHz, CDCl3) δ 7.66 (d, J = 7.5 Hz, 2H), 7.53 (t, J = 6.6 Hz, 2H), 7.33–7.26 (m, 2H), 7.20 (tt, J = 7.4, 0.9 Hz, 2H), 5.81 (ddt, J = 16.1, 11.0, 5.7 Hz, 1H), 5.23 (dd, J = 17.3, 1.6 Hz, 1H), 5.14 (dd, J = 10.4, 1.1 Hz, 1H), 4.54 (d, J = 5.8 Hz, 2H), 4.13 (t, J = 7.2 Hz, 1H), 4.01 (q, J = 7.1 Hz, 2H), 3.95 (q, J = 6.0 Hz, 1H), 3.25–3.03 (m, 1H), 2.27–2.12 (m, 2H), 1.88–1.60 (m, 1H), 1.53–1.34 (m, 2H), 1.12–1.05 (m, 12H), 0.84–0.70 (m, 4H). 13C NMR (101 MHz, CDCl3) δ 162.44, 141.17, 131.57, 127.60, 126.97, 119.85, 118.60, 73.89, 65.72, 64.80, 60.23, 53.84 (d, J = 5.1 Hz), 47.08, 45.71, 38.48, 36.34, 31.27, 28.96 (d, J = 6.9 Hz), 28.27, 22.82 (d, J = 1.8 Hz), 22.62, 22.54, 20.90, 14.06 (d, J = 7.3 Hz). MS calcd for C32H43N2O6 [M + H]+, 552.3; found 552.3.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70) to afford Fmoc-Lys(bhb)-OH (166 mg, 0.3 mmol, 57%). 1H NMR (400 MHz, CDCl3) δ 7.74 (d, J = 7.5 Hz, 2H), 7.49–7.43 (m, 2H), 7.37 (t, J = 7.4 Hz, 1H), 7.31–7.25 (m, 2H), 4.44–4.29 (m, 1H), 4.19 (t, J = 6.9 Hz, 1H), 4.12 (q, J = 7.1 Hz, 2H), 4.04 (td, J = 6.3, 4.8 Hz, 1H), 3.39–3.10 (m, 1H), 2.39–2.24 (m, 1H), 1.25 (t, J = 7.1 Hz, 3H), 1.17 (d, J = 6.1 Hz, 12H), 0.88 (t, J = 7.1 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 172.17, 171.20, 141.26, 132.24 (d, J = 2.9 Hz), 132.19, 132.09, 128.68, 128.55, 127.66, 127.07, 125.18, 119.93, 65.03, 60.41, 47.16 (d, J = 1.8 Hz), 45.39, 39.06, 28.32 (d, J = 2.2 Hz), 22.88 (d, J = 2.5 Hz), 21.03, 14.19. MS calcd for C29H39N2O6 [M + H]+, 511.3; found 511.3.
70) to afford Fmoc-Lys(bhb)-OH (166 mg, 0.3 mmol, 57%). 1H NMR (400 MHz, CDCl3) δ 7.74 (d, J = 7.5 Hz, 2H), 7.49–7.43 (m, 2H), 7.37 (t, J = 7.4 Hz, 1H), 7.31–7.25 (m, 2H), 4.44–4.29 (m, 1H), 4.19 (t, J = 6.9 Hz, 1H), 4.12 (q, J = 7.1 Hz, 2H), 4.04 (td, J = 6.3, 4.8 Hz, 1H), 3.39–3.10 (m, 1H), 2.39–2.24 (m, 1H), 1.25 (t, J = 7.1 Hz, 3H), 1.17 (d, J = 6.1 Hz, 12H), 0.88 (t, J = 7.1 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 172.17, 171.20, 141.26, 132.24 (d, J = 2.9 Hz), 132.19, 132.09, 128.68, 128.55, 127.66, 127.07, 125.18, 119.93, 65.03, 60.41, 47.16 (d, J = 1.8 Hz), 45.39, 39.06, 28.32 (d, J = 2.2 Hz), 22.88 (d, J = 2.5 Hz), 21.03, 14.19. MS calcd for C29H39N2O6 [M + H]+, 511.3; found 511.3.
(S)-2-(tert-butoxy)propanamide (mixture of stereoisomers). N-vinyl acetamides were synthesized according to published procedures.64 (S)-2-(tert-butoxy)propanoic acid (172 mg, 1.2 mmol) was dissolved in THF and cooled down to 0 °C and ethyl chloroformate (147 µL, 1.4 mmol, 1.4 eq.) and TEA (491 µL, 3.6 mmol, 3 eq.) were added and the reaction was left stirring for 30 minutes. Then, a 1 M aqueous NH4Cl (1.8 mL, 1.8 mmol, 1.5 eq.) was added to the reaction mixture and stirred for an additional 30 minutes at 0 °C. The reaction mixture was diluted with water and extracted with DCM three times, dried over MgSO4, filtered and concentrated in vacuo. The crude product was purified by flash column chromatography on silica gel (hexanes/EtOAc, 40
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) to afford (S)-2-(tert-butoxy)propanamide (160 mg, 1.1 mmol, 94%). 1H NMR (400 MHz, CDCl3) δ 3.93 (q, J = 6.9 Hz, 1H), 1.27 (d, J = 6.9 Hz, 3H), 1.15 (s, 9H). MS calcd for C7H16NO2 [M + H]+, 146.1; found 146.1.
60) to afford (S)-2-(tert-butoxy)propanamide (160 mg, 1.1 mmol, 94%). 1H NMR (400 MHz, CDCl3) δ 3.93 (q, J = 6.9 Hz, 1H), 1.27 (d, J = 6.9 Hz, 3H), 1.15 (s, 9H). MS calcd for C7H16NO2 [M + H]+, 146.1; found 146.1.
(S)-2-(tert-butoxy)-N-vinylpropanamide (mixture of stereoisomers). (S)-2-(tert-butoxy)propanamide (172 mg, 1.2 mmol), Cs2CO3 (384 mg, 1.4 mmol, 1.2 eq.) and CuI (18.6 mg, 0.1 mmol, 0.1 eq.) were dissolved in THF and flushed with nitrogen for 10 minutes. Vinyl iodide (174 µL, 1.2 mmol, 1 eq.) and DMEDA (52 µL mL, 0.6 mmol, 0.5 eq.) were added and the reaction was stirred under reflux for 16 hours. The crude reaction mixture was filtered through silica gel using EtOAc and concentrated in vacuo to afford (S)-2-(tert-butoxy)-N-vinylpropanamide (97 mg, 0.6 mmol, 47%) which was used without further purification. MS calcd for C9H18NO2 [M + H]+, 172.1; found 172.1.
3-(tert-butoxy)butanamide (mixture of stereoisomers). 3-(tert-butoxy)butanoic acid (78 mg, 0.4 mmol) was dissolved in THF and cooled down to 0 °C and ethyl chloroformate (65 µL, 0.6 mmol, 1.4 eq.) and TEA (204 µL, 1.2 mmol, 3 eq.) were added and the reaction was left stirring for 30 minutes. Then, a 1 M aqueous NH4Cl (735 µL, 0.8 mmol, 1.5 eq.) was added to the reaction mixture and stirred for an additional 30 minutes at 0 °C. The reaction mixture was diluted with water and extracted with DCM three times, dried over MgSO4, filtered and concentrated in vacuo. The crude product was purified by flash column chromatography on silica gel (hexanes/EtOAc, 40
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) to afford 3-(tert-butoxy)butanoic acid (64 mg, 0.3 mmol, 83%). 1H NMR (400 MHz, CDCl3) δ 4.32–4.27 (m, 2H), 2.37 (dd, J = 14.4, 4.9 Hz, 1H), 2.28 (ddd, J = 14.4, 5.6, 0.8 Hz, 1H), 1.23 (d, J = 7.1 Hz, 3H), 1.18 (s, 9H). MS calcd for C8H17NO2 [M + H]+, 160.1; found 160.1.
60) to afford 3-(tert-butoxy)butanoic acid (64 mg, 0.3 mmol, 83%). 1H NMR (400 MHz, CDCl3) δ 4.32–4.27 (m, 2H), 2.37 (dd, J = 14.4, 4.9 Hz, 1H), 2.28 (ddd, J = 14.4, 5.6, 0.8 Hz, 1H), 1.23 (d, J = 7.1 Hz, 3H), 1.18 (s, 9H). MS calcd for C8H17NO2 [M + H]+, 160.1; found 160.1.
3-(tert-butoxy)-N-vinylbutanamide (mixture of stereoisomers). 3-(tert-butoxy)butanamide (64 mg, 0.3 mmol), Cs2CO3 (96 mg, 0.4 mmol, 1.2 eq.) and CuI (4.7 mg, 0.03 mmol, 0.1 eq.) were dissolved in THF and flushed with nitrogen for 10 minutes. Vinyl iodide (44 µL, 0.3 mmol, 1 eq.) and DMEDA (13 µL, 0.15 mmol, 0.5 eq.) were added and the reaction was stirred under reflux for 16 hours. The crude reaction mixture was filtered through silica gel using EtOAc and concentrated in vacuo to afford 3-(tert-butoxy)-N-vinylbutanamide (40 mg, 0.2 mmol, 72%) which was used without further purification. MS calcd for C10H19NO2 [M + H]+, 186.1; found 186.2.
N-(2-Chloroethyl)-N-methylacetamide. 2-N-Methylaminoethyl chloride hydrochloride (100 mg, 0.77 mmol) was dissolved in acetic anhydride and pyridine (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and left stirring at room temperature for 2 hours. The reaction mixture was diluted with DCM and concentrated in vacuo by repeated coevaporation with toluene to afford N-(2-chloroethyl)-N-methylacetamide (140 mg, 0.76 mmol, 99%). MS calcd for C5H12ClNO [M + H]+, 136.1; found 136.1.
1, v/v) and left stirring at room temperature for 2 hours. The reaction mixture was diluted with DCM and concentrated in vacuo by repeated coevaporation with toluene to afford N-(2-chloroethyl)-N-methylacetamide (140 mg, 0.76 mmol, 99%). MS calcd for C5H12ClNO [M + H]+, 136.1; found 136.1.
2-Chloroacetimidamide. Using previously published procedures,31 2-chloroethanimidoate hydrochloride (250 mg, 1.9 mmol) was added to a stirring solution of sodium ethoxide (215 mg, 2.9 mmol, 1.5 eq.) in ethanol under a nitrogen atmosphere and stirred for 3 hours. Then, ammonium chloride (125 mg, 2.9 mmol, 1.5 eq.) was added in portions over 10 minutes and subsequently stirred for 24 hours. The reaction mixture was acidified with 4 M HCl in dioxane to pH 2 and concentrated in vacuo. Next, diethyl ether was added and the precipitates were filtered and dried to yield 2-chloroacetimidamide (91 mg, 1.0 mmol, 53% yield). MS calcd for C2H6ClN2 [M + H]+, 93.0; found 93.1.
Author contributions
Jordi C. J. Hintzen: conceptualization, methodology, formal analysis, investigation, validation, resources, writing – original draft. Kamiel D. Beckley: investigation, methodology, resources, writing – review & editing. Emily L. Goldberg: conceptualization, writing – review & editing. George M. Burslem: conceptualization, methodology, writing – review & editing, resources, supervision, funding acquisition.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: additional materials and methods, SI Fig. 1–3, copies of NMR spectra (PDF). See DOI: https://doi.org/10.1039/d5sc08759g.Acknowledgements
We thank members of the G. M. B. and E. L. G. labs for comments and intellectual discussion. We thank Juana G. Stollmaier and David W. Christianson for their kind donation of the plasmid encoding the catalytic domain of HDAC6. The E. L. G. lab is funded by The Shurl and Kay Curci Foundation, the Chan Zuckerberg Biohub; and the Sandler Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation (to E. L. G.). The G. M. B. lab is funded by the National Institute of General Medical Sciences (R35GM142505) to G.M.B.References
- A. S. Venne, L. Kollipara and R. P. Zahedi, Proteomics, 2014, 14, 513–524 CrossRef CAS PubMed.
- Y. L. Deribe, T. Pawson and I. Dikic, Nat. Struct. Mol. Biol., 2010, 17, 666–672 CrossRef CAS.
- G. Millán-Zambrano, A. Burton, A. J. Bannister and R. Schneider, Nat. Rev. Genet., 2022, 23, 563–580 CrossRef.
- J. M. Lee, H. M. Hammarén, M. M. Savitski and S. H. Baek, Nat. Commun., 2023, 14, 201 CrossRef CAS.
- J. Liu, C. Qian and X. Cao, Immunity, 2016, 45, 15–30 CrossRef CAS.
- C. E. W. Crawford and G. M. Burslem, Trends Cancer, 2025, 11(4), 403–420 CrossRef CAS PubMed.
- J. N. Beyer, N. R. Raniszewski and G. M. Burslem, ChemBioChem, 2021, 22, 17–42 CrossRef CAS PubMed.
- G. M. Burslem, Biochim. Biophys. Act. Gen. Sub., 2022, 1866, 130079 CrossRef CAS PubMed.
- R. Gupta, M. Sahu, D. Srivastava, S. Tiwari, R. K. Ambasta and P. Kumar, Ageing Res. Rev., 2021, 68, 101336 CrossRef CAS PubMed.
- C. L. Russell, S. Koncarevic and M. A. Ward, J. Alzheimer's Dis., 2014, 41, 345–364 CAS.
- B. S. Sharma, V. Prabhakaran, A. P. Desai, J. Bajpai, R. J. Verma and P. K. Swain, Oncogen, 2019, 2, 12 CrossRef.
- X. Wu, M. Xu, M. Geng, S. Chen, P. J. Little, S. Xu and J. Weng, Signal Transduct. Target. Ther., 2023, 8, 220 CrossRef CAS.
- M. M. Müller and T. W. Muir, Chem. Rev., 2015, 115, 2296–2349 CrossRef.
- B. C. Taylor and N. L. Young, Biochem. J., 2021, 478, 511–532 CrossRef CAS PubMed.
- N. N. Aye-Han, Q. Ni and J. Zhang, Curr. Opin. Chem. Biol., 2009, 13, 392–397 CrossRef CAS.
- J. C. J. Hintzen, H. Abujubara, D. Tietze and A. A. Tietze, Chem. Eur. J., 2024, 30, e202401103 CrossRef CAS PubMed.
- M. C. Araujo, R. I. Melo, E. Del Nery, M. F. Alves, M. A. Juliano, D. E. Casarini, L. Juliano and A. K. Carmona, J. Hypertens., 1999, 17, 665–672 CrossRef CAS.
- A. Caporale, F. Mascanzoni, B. Farina, M. Sturlese, G. Di Sorbo, R. Fattorusso, M. Ruvo and N. Doti, J. Biomol. Screen., 2016, 21, 701–712 CrossRef CAS.
- A. H. Simon, S. Liebscher, T. H. Aumüller, D. Treblow and F. Bordusa, Front. Microbiol., 2019, 10, 711 CrossRef PubMed.
- P. Kasperkiewicz, Front. Chem., 2021, 9, 639410 CrossRef CAS.
- H. Angliker, U. Neumann, S. S. Molloy and G. Thomas, Anal. Biochem., 1995, 224, 409–412 CrossRef CAS PubMed.
- M. Zessin, M. Meleshin, Z. Simic, D. Kalbas, M. Arbach, P. Gebhardt, J. Melesina, S. Liebscher, F. Bordusa, W. Sippl, C. Barinka and M. Schutkowski, Bioorg. Chem., 2021, 117, 105425 CrossRef CAS PubMed.
- M. Zessin, Z. Kutil, M. Meleshin, Z. Nováková, E. Ghazy, D. Kalbas, M. Marek, C. Romier, W. Sippl, C. Bařinka and M. Schutkowski, Biochemistry, 2019, 58, 4777–4789 CrossRef CAS PubMed.
- A. Yaron, A. Carmel and E. Katchalski-Katzir, Anal. Biochem., 1979, 95, 228–235 CrossRef CAS.
- H. K. Hustoft, H. Malerod, S. R. Wilson, L. Reubsaet, E. Lundanes and T. Greibrokk, Integr. Proteomics, 2012, 73, 74–92 Search PubMed.
- Z. A. Wang and P. A. Cole, Cell Chem. BIol., 2020, 27, 953–969 CrossRef CAS.
- J. Fuhrmann, K. W. Clancy and P. R. Thompson, Chem. Rev., 2015, 115, 5413–5461 CrossRef CAS PubMed.
- D. J. Slade, V. Subramanian, J. Fuhrmann and P. R. Thompson, Biopolymers, 2014, 101, 133–143 CrossRef CAS PubMed.
- J. C. J. Hintzen and J. Mecinović, Tetrahedron Lett., 2023, 124, 154602 CrossRef CAS.
- K. Arita, T. Shimizu, H. Hashimoto, Y. Hidaka, M. Yamada and M. Sato, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5291–5296 CrossRef CAS.
- S. Ofori, H. S. Desai, F. Shikwana, L. M. Boatner, E. R. Dominguez Iii, J. O. Castellón and K. M. Backus, Chem. Commun., 2024, 60, 8856–8859 RSC.
- M. D. Simon, F. Chu, L. R. Racki, C. C. de la Cruz, A. L. Burlingame, B. Panning, G. J. Narlikar and K. M. Shokat, Cell, 2007, 128, 1003–1012 CrossRef CAS PubMed.
- G. Proietti, G. Rainone, J. C. J. Hintzen and J. Mecinović, Bioconjugate Chem.br, 2020, 31, 844–851 CrossRef CAS PubMed.
- Q. Shu, J. Fan, B. Wang and J. Shi, Tetrahedron Lett., 2024, 135, 154888 CrossRef CAS.
- Y. Jing, Z. Liu, G. Tian, X. Bao, T. Ishibashi and X. D. Li, Cell Chem. Biol., 2018, 25, 166–174 CrossRef CAS PubMed.
- F. Li, A. Allahverdi, R. Yang, G. B. J. Lua, X. Zhang, Y. Cao, N. Korolev, L. Nordenskiöld and C. F. Liu, Angew. Chem., Int. Ed., 2011, 41, 9611–9614 CrossRef PubMed.
- Z. Fan, Z. Liu, N. Zhang, W. Wei, K. Cheng, H. Sun and Q. Hao, iScience, 2023, 26, 107757 CrossRef CAS PubMed.
- X. Zhang, R. Cao, J. Niu, S. Yang, H. Ma, S. Zhao and H. Li, Cell Discov., 2019, 5, 35 CrossRef.
- S. M. Abmayr and J. L. Workman, Cell Res., 2019, 29, 694–695 CrossRef.
- R. Du, Y. Gao, C. Yan, X. Ren, S. Qi, G. Liu, X. Guo, X. Song, H. Wang and J. Rao, iScience, 2024, 27, 110911 CrossRef CAS PubMed.
- W. C. Hallows, S. Lee and J. M. Denu, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10230–10235 CrossRef CAS PubMed.
- A. J. de Ruijter, A. H. van Gennip, H. N. Caron, S. Kemp and A. B. van Kuilenburg, Biochem. J., 2003, 370, 737–749 CrossRef CAS PubMed.
- C. Moreno-Yruela, D. Zhang, W. Wei, M. Bæk, W. Liu, J. Gao, D. Danková, A. L. Nielsen, J. E. Bolding and L. Yang, Sci. Adv., 2022, 8, eabi6696 CrossRef CAS.
- M. B. Gonzatti, J. C. J. Hintzen, I. Sharma, M. A. Najar, T. Tsusaka, M. M. Marcinkiewicz, C. V. D. S. Crispim, N. W. Snyder, G. M. Burslem and E. L. Goldberg, J. Biol. Chem., 2025, 110602 CrossRef CAS PubMed.
- T. Tsusaka, M. A. Najar, B. Schwarz, E. Bohrnsen, J. A. Oses-Prieto, H. Neudorf, C. Lee, J. P. Little, A. L. Burlingame, C. M. Bosio, G. M. Burslem and E. L. Goldberg, Nat. Chem. Biol., 2025, 21, 1387–1396 CrossRef CAS PubMed.
- S. Sun, Z. Xu, L. He, Y. Shen, Y. Yan, X. Lv, X. Zhu, W. Li, W.-Y. Tian and Y. Zheng, Nat. Commun., 2024, 15, 8377 CrossRef.
- R. Fellows, J. Denizot, C. Stellato, A. Cuomo, P. Jain, E. Stoyanova, S. Balázsi, Z. Hajnády, A. Liebert, J. Kazakevych, H. Blackburn, R. O. Corrêa, J. L. Fachi, F. T. Sato, W. R. Ribeiro, C. M. Ferreira, H. Perée, M. Spagnuolo, R. Mattiuz, C. Matolcsi, J. Guedes, J. Clark, M. Veldhoen, T. Bonaldi, M. A. R. Vinolo and P. Varga-Weisz, Nat. Commun., 2018, 9, 105 CrossRef PubMed.
- M. Huang, J. Zhang, C. Yan, X. Li, J. Zhang and R. Ling, Bioorg. Chem., 2019, 91, 103184 CrossRef CAS PubMed.
- C. Hubbert, A. Guardiola, R. Shao, Y. Kawaguchi, A. Ito, A. Nixon, M. Yoshida, X.-F. Wang and T.-P. Yao, Nature, 2002, 417, 455–458 CrossRef CAS.
- S. Kaur, P. Rajoria and M. Chopra, Cell. Oncol., 2022, 45, 779–829 CrossRef CAS.
- K. Sadoul and S. Khochbin, Biochem. J., 2016, 473, 1859–1868 CrossRef CAS PubMed.
- J. D. Osko and D. W. Christianson, Methods Enzymol., 2019, 626, 447–474 CAS.
- M. Bremang, A. Cuomo, A. M. Agresta, M. Stugiewicz, V. Spadotto and T. Bonaldi, Mol. BioSyst., 2013, 9, 2231–2247 RSC.
- R. Anand and R. Marmorstein, J. Biol. Chem., 2007, 282, 35425–35429 CrossRef CAS.
- C. Shiau, M. J. Trnka, A. Bozicevic, I. O. Torres, B. Al-Sady, A. L. Burlingame, G. J. Narlikar and D. G. Fujimori, Chem. Biol., 2013, 20, 494–499 CrossRef CAS PubMed.
- G. Proietti, G. Rainone, J. C. J. Hintzen and J. Mecinovic, Bioconjugate Chem., 2020, 31, 844–851 CrossRef CAS PubMed.
- A. H. K. Al Temimi, R. van der Wekken-de Bruijne, G. Proietti, H. Guo, P. Qian and J. Mecinovic, Bioconjugate Chem., 2019, 30, 1798–1804 CrossRef CAS PubMed.
- S. Mukherjee, K. A. Perez, C. Dubois, R. M. Nisbet, Q.-X. Li, S. Varghese, L. Jin, I. Birchall, V. A. Streltsov and L. J. Vella, ACS Chem. Neurosci., 2021, 12, 3719–3732 CrossRef CAS PubMed.
- W. J. Lu-Culligan, L. J. Connor, Y. Xie, B. E. Ekundayo, B. T. Rose, M. Machyna, A. P. Pintado-Urbanc, J. T. Zimmer, I. W. Vock, N. V. Bhanu, M. C. King, B. A. Garcia, F. Bleichert and M. D. Simon, Nature, 2023, 622, 173–179 CrossRef CAS PubMed.
- X.-J. Yang and E. Seto, Mol. Cell, 2008, 31, 449–461 CrossRef CAS PubMed.
- Z. Lu, Z. Cheng, Y. Zhao and S. L. Volchenboum, PloS one, 2011, 6, e28228 CrossRef CAS PubMed.
- V. Schwämmle, S. Sidoli, C. Ruminowicz, X. Wu, C.-F. Lee, K. Helin and O. N. Jensen, Mol. Cell. Proteomics, 2016, 15, 2715–2729 CrossRef PubMed.
- R. Collins and X. Cheng, Nuc. Acids Res., 2010, 38, 3503–3511 CrossRef CAS PubMed.
- T. Noguchi, M. Sekine, Y. Yokoo, S. Jung and N. Imai, Chem. Lett., 2013, 42, 580–582 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |