Open Access Article
Open Access ArticleSingle-cell structural lipidomics using a miniature dual-LIT mass spectrometer
Zhijun Cai,
Ningxi Li,
Simin Cheng,
Zheng Ouyang* and
Xiaoxiao Ma *
*
State Key Laboratory of Precision Measurement Technology and Instruments, Department of Precision Instruments, Tsinghua University, Beijing 100084, China. E-mail: ouyang@mail.tsinghua.edu.cn; maxx@mail.tsinghua.edu.cn
First published on 22nd December 2025
Abstract
Structural lipidomics provides comprehensive information on lipidomes, and there is great interest in applying it to single-cell analysis for accurate cell phenotyping and lipid pathway studies. However, structural lipidomics relies on tandem mass spectrometry analysis of a large number of lipid species, which remains challenging for single-cell analysis due to the small sample amounts. Herein, using a miniature dual-linear ion trap (LIT) mass spectrometer with enhanced ion processing capability, we developed an effective strategy to achieve annotations of more than 100 lipid species in a single cell with high structural specificity. The ion utilization efficiency was greatly improved with multi-stage MSn (n = 2–4) analysis performed for each lipid species to acquire structural information at different levels. From a single MDA-MB-468 cancer cell, we identified 100+ lipids, including 64 lipids at the acyl-chain sum composition level, 23 at the sn-position level, 30 at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location level, and 20 at the C
C location level, and 20 at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position level, primarily phosphatidylcholamines (PCs) and phosphatidylethanolamines (PEs). Significant variations in sn-position and C
C/sn-position level, primarily phosphatidylcholamines (PCs) and phosphatidylethanolamines (PEs). Significant variations in sn-position and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers were observed in doxorubicin-resistant and -sensitive K562 cells. With enriched information at the structural lipidomics level, the correlation was established between variations in lipidome and response to ferroptosis for human cancer cells.
C location isomers were observed in doxorubicin-resistant and -sensitive K562 cells. With enriched information at the structural lipidomics level, the correlation was established between variations in lipidome and response to ferroptosis for human cancer cells.
Introduction
Single-cell analysis has enabled and accelerated studies of cell-to-cell heterogeneities that are instrumental in understanding disease development.1–4 The technical advancement and applications of single-cell genomics, transcriptomics, and proteomics have made significant contributions to biological science. Lipidomics is an essential part of metabolomics that is also expected to provide important information regarding the biological processes of a single cell.5–9 Lipids serve as crucial cellular metabolites, constituting integral components of membranes and organelles and playing pivotal roles in processes such as cell division, endocytosis, protein export, and cell signaling.10–12 The combination of lipidomics with other omics methods applied to single-cell studies has contributed to the understanding of the genetic and molecular basis of diseases, molecular changes that occur during the development of organisms, and the immune system's response to infections.13,14 Mass spectrometry (MS) is the most effective tool for lipidomics due to its exceptional structural characterization capability.In the past decade, powerful lipid structural characterization tools have emerged that have unraveled the complexity of the human lipidome, in particular at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location/geometry and fatty acyl sn-position levels.15–17 These tools rely on tandem MS (MS/MS or MSn) analysis, and were also shown to be effective in combination with chemical derivatization,18,19 such as for PB-MS/MS (PB, Paternò–Büchi reaction20). Consequently, these technical advancements also led to the consensus that the field of lipidomics has entered the era of structural lipidomics. Alternative lipid metabolic pathways have been revealed,21,22 and potential disease biomarkers have been proposed,23,24 which are critical for understanding diseases and finding new therapeutic targets.
C location/geometry and fatty acyl sn-position levels.15–17 These tools rely on tandem MS (MS/MS or MSn) analysis, and were also shown to be effective in combination with chemical derivatization,18,19 such as for PB-MS/MS (PB, Paternò–Büchi reaction20). Consequently, these technical advancements also led to the consensus that the field of lipidomics has entered the era of structural lipidomics. Alternative lipid metabolic pathways have been revealed,21,22 and potential disease biomarkers have been proposed,23,24 which are critical for understanding diseases and finding new therapeutic targets.
The application of structural lipidomic technologies for single-cell analysis, however, faces significant challenges stemming from the limited sample availability of a single cell, especially for mammalian cells.25 Conventional mass spectrometers with MS/MS capabilities are mostly designed for coupling with liquid chromatography, where ions from a sample are not utilized at high efficiency when MS/MS analysis is performed. Using a commercial ion trap, triple-quadrupole, or quadrupole-time-of-flight (TOF) mass spectrometers as an example, ions of only one target precursor are used for each MS/MS scan, with all the others wasted.26 The consequence of using these instruments for single-cell structural lipidomics analysis is low coverage of lipid species that can be identified with a high level of structural confidence.
Numerous methods have been explored to address these limitations for single-cell analysis. Qin et al. developed ID-organic cytoMS, incorporating an efficient online lysis system that extended single-cell analysis time from seconds to tens of seconds.27 This approach enabled the identification of 150 metabolite species from a single MCF-7 cancer cell, although the number of identified lipid species remained lower, likely due to sample dilution during analysis. The Li group extended trapped ion mobility separation to dual-polarity ionization mass spectrometry imaging for in situ single-cell lipidomics analysis. With this method, it was revealed that a total of 185 lipids were derived from single cells,28 but lipids are identified at the lipid sum composition level without in-depth structural information such as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location and sn-positions of fatty acyls. We explored the combination of on-demand nano-electrospray ionization (nanoESI) with photochemical derivatization using the PB reaction,29 empowering single-cell lipid analysis with C
C location and sn-positions of fatty acyls. We explored the combination of on-demand nano-electrospray ionization (nanoESI) with photochemical derivatization using the PB reaction,29 empowering single-cell lipid analysis with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location specificity. However, the coverage of the lipid species was still limited by the number of MS/MS analyses and ultimately the sample amount from a single cell.
C location specificity. However, the coverage of the lipid species was still limited by the number of MS/MS analyses and ultimately the sample amount from a single cell.
To truly apply structural lipidomics for single-cell analysis, it is highly desirable to increase the coverage of the lipid species from a single cell, which indicates that MSn analysis with significantly improved efficiency for sample usage is required. It should be noted that MS3 or MS4 might also be required for comprehensive lipid structural characterization. For instance, determining the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location in an sn-specific fatty acyl requires two rounds (sn-1 and sn-2) of MS4 analysis for ultimate structure determination.30,31
C location in an sn-specific fatty acyl requires two rounds (sn-1 and sn-2) of MS4 analysis for ultimate structure determination.30,31
With these considerations, we explored the development of a high-coverage single-cell structural lipidomics method utilizing a miniature dual-linear ion trap (LIT) mass spectrometer.32,33 Ions were stored in LIT 1 and could be partially, mass-selectively transferred to LIT 2 for MSn (n = 2–4) analysis. That facilitated the comprehensive and targeted interrogation of the lipidome. The single-cell structural lipidomics approach enabled the identification of approximately 100 lipids from a single MDA-MB-468 breast cancer cell, including the precise determination of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations in sn-specific fatty acyls of phospholipids, representing the most in-depth characterization of phospholipids within single cells to date. Although our method is highly effective for PCs and PEs, its application to other lipid classes may require further optimization.
C locations in sn-specific fatty acyls of phospholipids, representing the most in-depth characterization of phospholipids within single cells to date. Although our method is highly effective for PCs and PEs, its application to other lipid classes may require further optimization.
We successfully applied the method to distinguish between doxorubicin (DOX)-resistant and DOX-sensitive human chronic myelogenous leukemia (CML) K562 cells based on their unique lipidomic profiles, leveraging the quantitative information obtained at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location and sn-position levels. We also evaluated the erastin-induced ferroptosis in human breast and liver cancer cell lines and uncovered a transition state between normal and ferroptotic cells. These findings underscore the significant potential of this novel single-cell structural lipidomics method for advancing the understanding of cellular heterogeneity.
C location and sn-position levels. We also evaluated the erastin-induced ferroptosis in human breast and liver cancer cell lines and uncovered a transition state between normal and ferroptotic cells. These findings underscore the significant potential of this novel single-cell structural lipidomics method for advancing the understanding of cellular heterogeneity.
Results and discussion
The single-cell MS platform based on dual-LIT MS
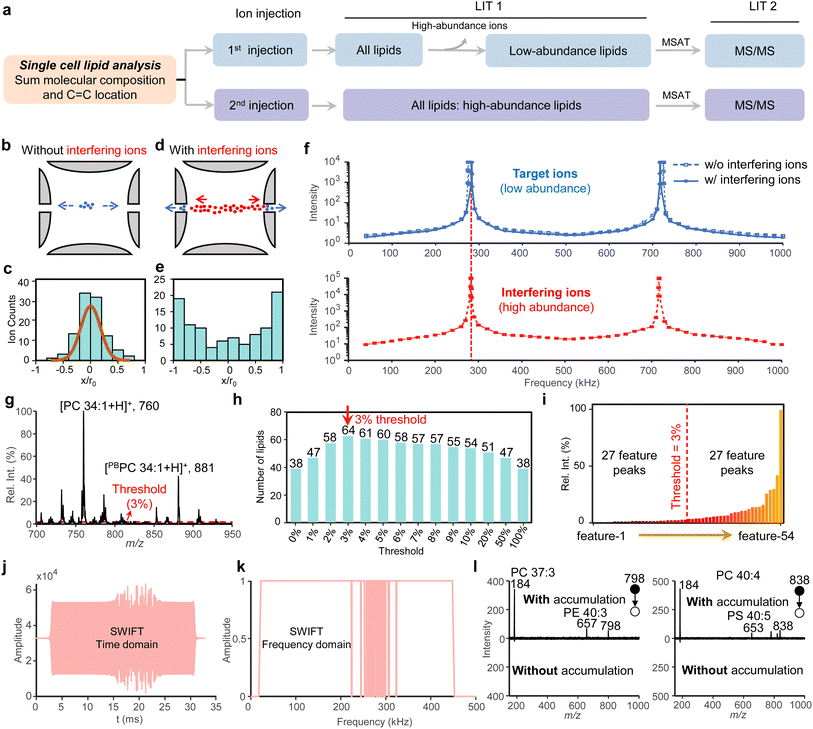
The platform integrated the single-cell ion source with the dual-LIT MS platform (Fig. 1a). The single-cell sampling and ionization method was modified from the reported methods that involved cell fixation, migration, derivatization, and ionization.29 The optimized workflow is shown in Fig. S1a and comprises three steps. Step 1: single cells were suspended in a methanol/H2O mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and migrated to the capillary tip via gravity, followed by drying. This solvent condition was chosen to minimize lipid dissolution in the solvent and promote on-demand ionization for sensitive analysis. Step 2: lipids were derivatized using 2-acetylpyridine (2-AP, 121.14 g mol−1) in acetonitrile/water (1
1, v/v) and migrated to the capillary tip via gravity, followed by drying. This solvent condition was chosen to minimize lipid dissolution in the solvent and promote on-demand ionization for sensitive analysis. Step 2: lipids were derivatized using 2-acetylpyridine (2-AP, 121.14 g mol−1) in acetonitrile/water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) via a droplet-assisted method. Step 3: lipids were ionized using on-demand nanoESI with methanol/acetonitrile (1
1, v/v) via a droplet-assisted method. Step 3: lipids were ionized using on-demand nanoESI with methanol/acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) containing 1% formic acid.34 This gravity-based cell migration method, different from electro-migration, enabled parallel processing of multiple cells (>100) with an average processing time of less than 10 s per cell. Furthermore, the methanol/H2O-based cell suspension method eliminated the need for glutaraldehyde fixation, preserving crucial phospholipid species such as PEs (Fig. S1c). To ensure optimal performance, the time for matching the single-cell ion source with dual-LIT MS was carefully optimized (Fig. S2). However, the round of sampling from a single cell using on-demand nanoESI was limited to four, which was fewer than achievable with glutaraldehyde fixation methods,29 as shown in Fig. S3. This limitation might be attributed to potential lipid loss during the methanol/H2O-based cell suspension process.
1, v/v) containing 1% formic acid.34 This gravity-based cell migration method, different from electro-migration, enabled parallel processing of multiple cells (>100) with an average processing time of less than 10 s per cell. Furthermore, the methanol/H2O-based cell suspension method eliminated the need for glutaraldehyde fixation, preserving crucial phospholipid species such as PEs (Fig. S1c). To ensure optimal performance, the time for matching the single-cell ion source with dual-LIT MS was carefully optimized (Fig. S2). However, the round of sampling from a single cell using on-demand nanoESI was limited to four, which was fewer than achievable with glutaraldehyde fixation methods,29 as shown in Fig. S3. This limitation might be attributed to potential lipid loss during the methanol/H2O-based cell suspension process.
The dual-LIT MS system, described in detail in the ‘Dual-LIT MS System’ section of the SI, played a crucial role in this study. Ionized lipids from single cells were trapped in LIT 1 and mass-selectively transferred to LIT 2 for MSn (n = 2–4) analysis (Fig. 1a). Optimization of the dual-LIT MS system, particularly the vacuum regulation system (Fig. S4a), significantly enhanced ion transfer efficiency and storage time, leading to improved sensitivity and enabling multiple ion analysis capabilities.35 Notably, dual-LIT MS efficiently stored ions for more than 30 s with minimal ion loss (<10%) (Fig. S4b–d), and therefore, it is well-suited for single-cell lipidomics by maximizing ion utilization. This platform also offers versatile MSn capabilities tailored for comprehensive lipid structure characterization.
Three unique operational modes were employed in this work (Fig. 1b): (i) mass-selective axial transfer (MSAT), enabling sequential and selective ion transfer from LIT 1 to LIT 2 for efficient multiple MS/MS analyses with high ion utilization;33 (ii) multiple mass-selective partial transfers (MMPTs), facilitating the transfer of ions of a specific m/z in multiple batches to acquire detailed structural information at various levels; (iii) selective accumulation of low-abundance ions, which enhanced the detection of low-abundance ions by accumulating them in LIT 1 via ejecting high-abundance ions. The MMPT was developed based on the MSAT method. MSAT utilized an AC signal with a sufficiently large amplitude to efficiently transfer ions from LIT 1 to LIT 2. By reducing the AC amplitude, only a portion of the ions was transferred (Fig. S5). This principle formed the basis of MMPTs, where partial ion transmission was repeated multiple times until all ions of a target lipid were transferred for subsequent MSn analysis (Fig. S6 and 7).
The dual-LIT MS platform features unique ion manipulation capabilities, including the ability to perform MSn analysis and three operational modes. Leveraging the optimized vacuum system, the platform exhibits exceptional ion storage capacity, maximizing ion utilization. In conjunction with optimized single-cell sampling and ionization methods, this integrated single-cell MS platform enables comprehensive and detailed lipid structural characterization at the omics level. The lipid structures characterized include lipid acyl-chain sum compositions, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations, sn-positions of fatty acyl, and C
C locations, sn-positions of fatty acyl, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations in sn-specific fatty acyl (C
C locations in sn-specific fatty acyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position).
C/sn-position).
High coverage lipid analysis with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) C location specificity
C location specificity
The dual-LIT mass spectrometer can maximize ion utilization due to its excellent ion storage capability. However, achieving high-coverage lipid profiling within single cells remains challenging. During MSAT, high-abundance ions could interfere with the transmission of low-abundance ions, particularly those with m/z values differing by only 2 Da, as evidenced in Fig. S8. Simulation studies, detailed in the ‘Theoretical model and simulation method’ section of the SI, elucidated the mechanism of the interference. When only target ions (blue) were present in the ion trap, they exhibited stable motion (Fig. 2b and c). However, the introduction of high-abundance interfering ions (red) disrupted this stability (Fig. 2d and e).
Analysis of ion movement frequencies revealed that the resonance point of the target ion was shifted towards the resonance point of the interfering ion (Fig. 2f). The frequency shift could be attributed to the collective interactions of ions, also known as the space-charge effect.36,37 The inherent characteristics of cellular lipidomes further exacerbated the interference between different ions. On the one hand, lipid ion abundances within a single cell could span several orders of magnitude. On the other hand, the presence of phospholipids with varying degrees of unsaturation resulted in closely spaced m/z values, often differing by only 2 Da, which significantly complicates ion isolation and transmission.
To enhance the dynamic range for lipid detection, a targeted lipidomic strategy was developed, as illustrated in Fig. 2a, to enable high coverage and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C structural specificity of lipid analysis within single cells. This strategy, implemented on the single-cell MS platform, involved the separate analysis of high- and low-abundance lipid ions to maximize single-cell lipidome coverage. Low-abundance ions were selectively accumulated in LIT 1, utilizing mode (iii) depicted in Fig. 1b, and subjected to multiple rounds of MS/MS. In contrast, high-abundance ions were analyzed following a separate ion injection. Accumulation of low-abundance ions was achieved using a stored waveform inverse Fourier transform (SWIFT) waveform.38,39 The SWIFT waveform, a short-duration (approximately 35 ms) broadband voltage signal (Fig. 2j), efficiently isolated ions of specific m/z values from a complex mixture. Secular frequencies of ions to be stored in LIT 1 were selectively excluded from the SWIFT waveform, resulting in ‘notches’ in the frequency spectrum (Fig. 2k). Of note, the average time required for ion isolation using SWIFT (radial ejection) was significantly shorter (<1 ms) than using axial ejection (MSAT) due to the considerably smaller radial dimension of the LIT compared to its axial length.
C structural specificity of lipid analysis within single cells. This strategy, implemented on the single-cell MS platform, involved the separate analysis of high- and low-abundance lipid ions to maximize single-cell lipidome coverage. Low-abundance ions were selectively accumulated in LIT 1, utilizing mode (iii) depicted in Fig. 1b, and subjected to multiple rounds of MS/MS. In contrast, high-abundance ions were analyzed following a separate ion injection. Accumulation of low-abundance ions was achieved using a stored waveform inverse Fourier transform (SWIFT) waveform.38,39 The SWIFT waveform, a short-duration (approximately 35 ms) broadband voltage signal (Fig. 2j), efficiently isolated ions of specific m/z values from a complex mixture. Secular frequencies of ions to be stored in LIT 1 were selectively excluded from the SWIFT waveform, resulting in ‘notches’ in the frequency spectrum (Fig. 2k). Of note, the average time required for ion isolation using SWIFT (radial ejection) was significantly shorter (<1 ms) than using axial ejection (MSAT) due to the considerably smaller radial dimension of the LIT compared to its axial length.
To implement the targeted lipidomics strategy outlined in Fig. 2a, it was necessary to first identify the target peaks for MS/MS analysis. A typical single-cell MS1 spectrum following PB reaction is shown in Fig. 2g. By repeatedly collecting lipid MS1 data from 10 single cells, 54 stable mass spectral features were identified for MDA-MB-468 cells (Fig. S9). Subsequently, the threshold for distinguishing high- and low-abundance ions was optimized. During the optimization process, 0.5 µL of cellular extracts in methanol/acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) was transferred to the nanoESI capillary to test repeatability. Lipids that were successfully identified in three replicate analyses were considered for further investigation. As illustrated in Fig. 2h, a 3% relative intensity (Rel. Int. (%)) threshold was determined to be the optimal threshold for distinguishing low-abundance ions from high-abundance ions. It should be noted that the 3% relative intensity threshold for distinguishing high- and low-abundance ions was optimized using the MDA-MB-468 cell line. While this threshold was also applicable to the MCF-7 and BT-474 breast cancer cells analyzed in this study, its universality may be limited for cell types with vastly different lipid compositions, and re-optimization would be necessary for such applications.
1, v/v) was transferred to the nanoESI capillary to test repeatability. Lipids that were successfully identified in three replicate analyses were considered for further investigation. As illustrated in Fig. 2h, a 3% relative intensity (Rel. Int. (%)) threshold was determined to be the optimal threshold for distinguishing low-abundance ions from high-abundance ions. It should be noted that the 3% relative intensity threshold for distinguishing high- and low-abundance ions was optimized using the MDA-MB-468 cell line. While this threshold was also applicable to the MCF-7 and BT-474 breast cancer cells analyzed in this study, its universality may be limited for cell types with vastly different lipid compositions, and re-optimization would be necessary for such applications.
In this study, we employed a relative quantification method, which was straightforward for quantifying the relative amounts of lipid isomers. Fig. 2i shows the distribution of high- and low-abundance ions. For the MDA-MB-468 cell, 27 low-abundance and 27 high-abundance lipid ions were detected. In particular, the SWIFT waveform (Fig. 2j and k) was generated according to the identified low-abundance feature peaks. The isolation efficiency of lipid targets using the SWIFT waveform exceeded 85%, as demonstrated in Fig. S10.
Following the establishment of the analysis strategy, multiple single cells were analyzed. The gas pressure curves for analyzing high- and low-abundance ions are presented in Fig. S11. The time for an injection was 100 ms, and an MSAT was 50 ms. Each MS/MS analysis required 900 ms for data acquisition. Consequently, the total analysis time for low- and high-abundance ions was approximately 50 s. The efficacy of ion accumulation was demonstrated by comparing MS/MS spectra of some specific low-abundance ions with and without accumulation (Fig. 2l). An improvement greater than 10-fold in the signal-to-noise (S/N) ratios of lipids was observed. This enhanced sensitivity enabled the identification of previously undetectable lipids, including PC 37:3, PE 40:3, PC 40:4, and PS 40:5, based on the presence of their characteristic fragment ions.
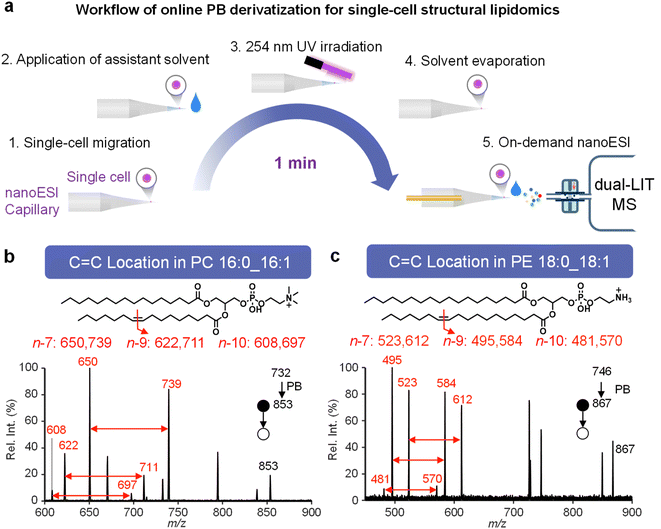
Among the 54 feature peaks identified in MDA-MB-468 cells, 9 were PB product ions, including PBPC 32:1, PBPC 34:2, PBPC 34:1, PBPC 36:3, PBPC 36:2, PBPC 36:1, PBPC 38:4, PBPE 36:2, and PBPE 36:1. By analyzing these PB product ions via MS/MS, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location of lipids can be determined. This is achieved through a single-cell online PB reaction process, which is illustrated in Fig. 3a and consists of five steps, all completed within approximately 1 minute: (1) single-cell migration to the capillary tip; (2) application of 10 mmol L−1 2-AP in acetonitrile/H2O (v/v, 1
C location of lipids can be determined. This is achieved through a single-cell online PB reaction process, which is illustrated in Fig. 3a and consists of five steps, all completed within approximately 1 minute: (1) single-cell migration to the capillary tip; (2) application of 10 mmol L−1 2-AP in acetonitrile/H2O (v/v, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); (3) 254 nm UV irradiation for 30 s; (4) solvent evaporation; (5) on-demand nanoESI analysis. Diagnostic ions for C
1); (3) 254 nm UV irradiation for 30 s; (4) solvent evaporation; (5) on-demand nanoESI analysis. Diagnostic ions for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations at n-10, n-9, and n-7 locations were detected in the MS/MS spectra of PBPC 32:1 and PBPE 36:1 (Fig. 3b and c). Additional MS/MS spectra of these PB product ions are presented in Fig. S13 and S14.
C locations at n-10, n-9, and n-7 locations were detected in the MS/MS spectra of PBPC 32:1 and PBPE 36:1 (Fig. 3b and c). Additional MS/MS spectra of these PB product ions are presented in Fig. S13 and S14.
All MS/MS spectra of the 54 feature peaks observed in the MDA-MB-468 cell are shown in Fig. S15. Peaks with an S/N ratio of >3 in the MS/MS spectra were considered for subsequent analysis. Lipid acyl-chain sum compositions and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations were determined based on the characteristic fragment ions generated during MS/MS analysis. For instance, after CID PCs generate characteristic fragments at m/z 184, PEs exhibit a characteristic neutral loss of 141 Da, and PSs exhibit a characteristic neutral loss of 185 Da. In total, 64 lipids were identified at the molecular sum composition level (Fig. S16 and Table S1), and 30 lipids were characterized at the C
C locations were determined based on the characteristic fragment ions generated during MS/MS analysis. For instance, after CID PCs generate characteristic fragments at m/z 184, PEs exhibit a characteristic neutral loss of 141 Da, and PSs exhibit a characteristic neutral loss of 185 Da. In total, 64 lipids were identified at the molecular sum composition level (Fig. S16 and Table S1), and 30 lipids were characterized at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location level from a single MDA-MB-468 cell (Table S2), demonstrating the method's ability to analyze lipids with high coverage and C
C location level from a single MDA-MB-468 cell (Table S2), demonstrating the method's ability to analyze lipids with high coverage and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C specificity. Of note, PCs and PEs are the primary lipids detected, and the lower detection of PS and SM species could be attributed to several factors, including their inherently lower ionization efficiency and the targeted analysis strategy, which prioritized ions based on pre-defined abundance thresholds.
C specificity. Of note, PCs and PEs are the primary lipids detected, and the lower detection of PS and SM species could be attributed to several factors, including their inherently lower ionization efficiency and the targeted analysis strategy, which prioritized ions based on pre-defined abundance thresholds.
sn-Position and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) C/sn-position specificity of lipid analysis
C/sn-position specificity of lipid analysis
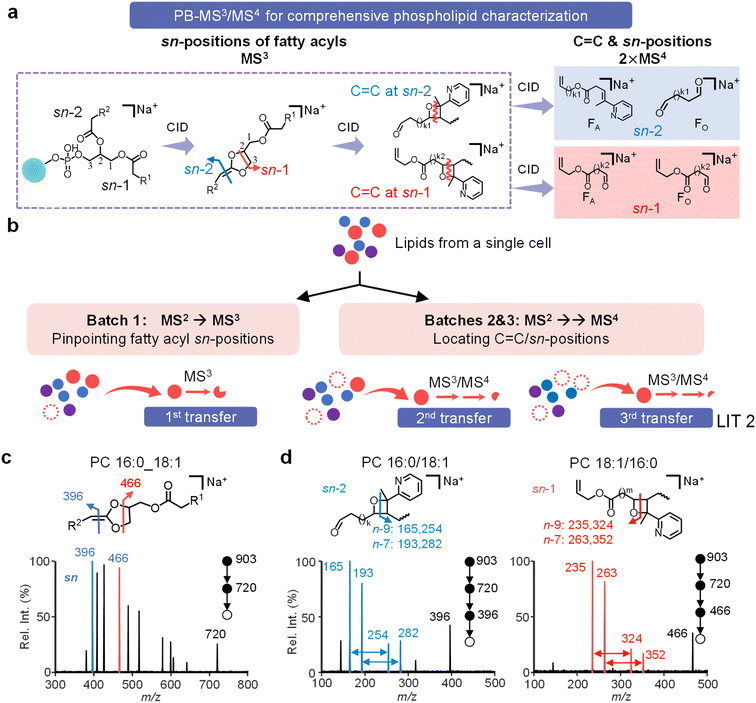
To achieve the ultimate lipid structural characterization for single-cell analysis, we implemented a strategy of combined MS3/MS4 analysis of sodiated PB products for the lipidome.31 MS3 analysis of these sodiated products releases sn-specific fatty acyls containing the 2-AP-derivatized C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond. Further MS4 analysis allows C
C bond. Further MS4 analysis allows C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location determination in the sn-specific fatty acyl, avoiding ambiguities in the determination of C
C location determination in the sn-specific fatty acyl, avoiding ambiguities in the determination of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location and sn-positions of fatty acyls for phospholipids (Fig. 4a).
C location and sn-positions of fatty acyls for phospholipids (Fig. 4a).
The PB-MS3/MS4 method requires a minimum of three MSn analyses to fully elucidate the complete structure of a target phospholipid. To evaluate the performance of this strategy, we performed MS3 analysis of mixed PC 16:0/18:1(n-9) and PC 18:1(n-9)/16:0 (Fig. S17), releasing sn-specific fatty acyls containing 2-AP derivatized C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (m/z 380/396 and 466 for C18:1 at sn-2 and sn-1, respectively). MS4 analysis of these sn-specific fragments confirmed the C
C (m/z 380/396 and 466 for C18:1 at sn-2 and sn-1, respectively). MS4 analysis of these sn-specific fragments confirmed the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location within sn-1 and sn-2 C18:1 fatty acyl, both at the n-9 location.
C location within sn-1 and sn-2 C18:1 fatty acyl, both at the n-9 location.
Using conventional MS instrumentation, without ion accumulation, a lipid precursor can only sustain one round of MS/MS, posing a challenge for comprehensive lipid characterization that requires multiple rounds of MSn analysis. The dual-LIT MS platform offers the flexibility to tackle this challenge via the MMPT mode, as shown in Fig. 1b. With the target lipid stored in LIT 1, MMPT enables partial transfers of these stored lipid ions to LIT 2 in a time sequence, enabling the detailed and complete MSn analysis required for ultimate phospholipid identification, and sn-position and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomer quantitation in a single-cell lipidome. Operationally, derivatized lipids were transferred from LIT 1 to LIT 2 in three batches and subjected to one MS3 and two MS4 analyses (Fig. 4b).
C location isomer quantitation in a single-cell lipidome. Operationally, derivatized lipids were transferred from LIT 1 to LIT 2 in three batches and subjected to one MS3 and two MS4 analyses (Fig. 4b).
To facilitate sodium-adduct formation, a spray solvent of methanol/acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), added to 100 µM sodium acetate, was used. Within LIT 1, 9 sodiated PB lipid products were accumulated after the removal of intact lipid ions in the low-mass range ( Fig. S18). Five of the most abundant derivatized lipids, including PBPC 32:1 (m/z 875), PBPC 34:2 (m/z 901), PBPC 34:1 (m/z 903), PBPC 36:2 (m/z 929), and PBPC 36:1 (m/z 931), were subjected to MSn analyses (one MS3 and two MS4) via MMPT. The remaining four lipids were analyzed by PB-MS3 for the determination of sn-positions of fatty acyls. Analysis of all these lipid products in an MDA-MB-468 cell was completed within approximately 25 s.
1, v/v), added to 100 µM sodium acetate, was used. Within LIT 1, 9 sodiated PB lipid products were accumulated after the removal of intact lipid ions in the low-mass range ( Fig. S18). Five of the most abundant derivatized lipids, including PBPC 32:1 (m/z 875), PBPC 34:2 (m/z 901), PBPC 34:1 (m/z 903), PBPC 36:2 (m/z 929), and PBPC 36:1 (m/z 931), were subjected to MSn analyses (one MS3 and two MS4) via MMPT. The remaining four lipids were analyzed by PB-MS3 for the determination of sn-positions of fatty acyls. Analysis of all these lipid products in an MDA-MB-468 cell was completed within approximately 25 s.
MS3 analysis of PBPC 34:1 (m/z 903) identified it as a mixture of PC 16:0/18:1 (m/z 396) and PC 18:1/16:0 (m/z 466) (Fig. 4c). The MS3 spectra of the other eight lipids are shown in Fig. S19. Subsequent MS4 analyses of the sn-specific fatty acyls (m/z 396, 466) showed the C18:1 fatty acyl at the sn-1 and sn-2 positions to be a mixture of n-9 and n-7 isomers, as evidenced by the corresponding pairs of diagnostic ions (highlighted in red in Fig. 4d). Our method revealed PC 16:0/18:1 to be composed of four distinct isomers in MDA-MB-468 cells, i.e., PC 16:0/18:1(n-9), PC 16:0/18:1(n-7), PC 18:1(n-9)/16:0, and PC 18:1(n-7)/16:0. These findings strongly support the method's capability for unprecedented single-cell lipidome analysis with high structural specificity.
Multi-dimensional in-depth single-cell lipidome characterization
The technique enables comprehensive lipid characterization at the single-cell level, encompassing lipid sum compositions, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations, sn-positions of fatty acyls, and C
C locations, sn-positions of fatty acyls, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-positions. Three rounds of sampling from a single cell were required for this comprehensive analysis. The first two rounds of sampling focused on determining lipid sum compositions and C
C/sn-positions. Three rounds of sampling from a single cell were required for this comprehensive analysis. The first two rounds of sampling focused on determining lipid sum compositions and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations. The third round of sampling was dedicated to identifying the sn-position and C
C locations. The third round of sampling was dedicated to identifying the sn-position and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position. From a single MDA-MB-468 cell, we successfully identified 64 molecular lipids (35 PCs, 20 PEs, 8 PSs, and 1 SM), 30 lipid C
C/sn-position. From a single MDA-MB-468 cell, we successfully identified 64 molecular lipids (35 PCs, 20 PEs, 8 PSs, and 1 SM), 30 lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers, and 23 lipid sn-position isomers (Table S3). Furthermore, 20 distinct lipids were characterized at C
C location isomers, and 23 lipid sn-position isomers (Table S3). Furthermore, 20 distinct lipids were characterized at C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position levels (Table S4).
C/sn-position levels (Table S4).
Excluding duplicated lipids, a total of 55 lipids were identified at the lipid sum composition level, with 16 identified at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location level, 10 lipids identified at the sn-position level, and 24 identified at the combined C
C location level, 10 lipids identified at the sn-position level, and 24 identified at the combined C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position level. In particular, a total of 8 isomers were identified for PC 34:2, including two isomers at the sn-position level (PC 16:0/18:2 (n-6, 9), and PC 18:2 (n-6, 9)/16:0) and six isomers at the C
C/sn-position level. In particular, a total of 8 isomers were identified for PC 34:2, including two isomers at the sn-position level (PC 16:0/18:2 (n-6, 9), and PC 18:2 (n-6, 9)/16:0) and six isomers at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position level (PC 16:1/18:1(n-7), PC 16:1/18:1(n-9), PC 18:1(n-7)/16:1, and PC 18:1(n-9)/16:1, PC 16:1/18:1(n-10), and PC 18:1(n-10)/16:1). These findings demonstrate the remarkable depth of lipid structural information that can be obtained from a single cell using our developed method.
C/sn-position level (PC 16:1/18:1(n-7), PC 16:1/18:1(n-9), PC 18:1(n-7)/16:1, and PC 18:1(n-9)/16:1, PC 16:1/18:1(n-10), and PC 18:1(n-10)/16:1). These findings demonstrate the remarkable depth of lipid structural information that can be obtained from a single cell using our developed method.
This single-cell lipidomics method was successfully applied to MCF-7 and BT-474 cell lines, and the identification results are summarized in Tables S5–10. A total of 60 molecular lipid species, 28 lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers, 21 lipid sn-position isomers, and 20 lipid C
C location isomers, 21 lipid sn-position isomers, and 20 lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position isomers were detected for MCF-7 cells. For BT-474 cells, a total of 56 molecular lipid species, 23 lipid C
C/sn-position isomers were detected for MCF-7 cells. For BT-474 cells, a total of 56 molecular lipid species, 23 lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers, 18 lipid sn-position isomers, and 20 lipid C
C location isomers, 18 lipid sn-position isomers, and 20 lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-position isomers were detected (Fig. S20). Distinct lipid profiles were acquired and can be used for the accurate grouping of MDA-MB-468, MCF-7, and BT-474 cells. Of note, the number of different lipids with structural specificity identified from a human breast cancer cell is higher than that which can be obtained with existing methods.27–29,40,41
C/sn-position isomers were detected (Fig. S20). Distinct lipid profiles were acquired and can be used for the accurate grouping of MDA-MB-468, MCF-7, and BT-474 cells. Of note, the number of different lipids with structural specificity identified from a human breast cancer cell is higher than that which can be obtained with existing methods.27–29,40,41
The quantification of lipid isomers in this study is based on the relative intensities of their diagnostic ions. This approach assumes that the ionization efficiencies of isomers derived from the same parent ion are comparable, and that diagnostic ions for different isomers are generated with similar fragmentation efficiencies. While these assumptions are necessary for relative quantification at the single-cell level, we acknowledge that they could be influenced by factors such as differential fragmentation or ion trap residence times. However, the high structural similarity of isomers, particularly for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers, suggests that these effects are likely minor. The consistency of our isomer ratio data with that in the existing literature further supports the validity of this quantitative approach.
C location isomers, suggests that these effects are likely minor. The consistency of our isomer ratio data with that in the existing literature further supports the validity of this quantitative approach.
In an ideal scenario, quantification could be achieved by spiking a known amount of an internal standard into a single cell. However, due to the extremely small volume of a single cell (<pL), it remains technically challenging to accurately introduce such a minute quantity. For the derivatization step, reproducibility at the single-cell level is achievable because the reaction is relatively stable. Quantification was achieved by normalizing lipid signals to either mutual internal standards (for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C isomers with similar efficiencies) or a single intense benchmark peak (e.g., m/z 760 for PC 34:1). Therefore, to ensure robust reproducibility, several strategies can be employed: applying internal calibration standards for signal correction; developing optimized normalization protocols, such as those utilizing stable isotope-labeled or endogenous housekeeping lipids, to correct for cell-to-cell heterogeneity; and integrating real-time monitoring systems for immediate quality assurance.
C isomers with similar efficiencies) or a single intense benchmark peak (e.g., m/z 760 for PC 34:1). Therefore, to ensure robust reproducibility, several strategies can be employed: applying internal calibration standards for signal correction; developing optimized normalization protocols, such as those utilizing stable isotope-labeled or endogenous housekeeping lipids, to correct for cell-to-cell heterogeneity; and integrating real-time monitoring systems for immediate quality assurance.
In addition to comprehensive single-cell lipidome analysis including lipid isomers, we next aimed to evaluate the performance of our method with respect to quantitative analysis of lipid isomers (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location, sn-position). Fig. 5a shows the distribution of C
C location, sn-position). Fig. 5a shows the distribution of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers (n-7, n-9, and n-10) for phospholipids containing a C18:1 and C16:1 fatty acyl. Notably, the proportion of C18:1 (n-7) is greater than 80% (calculated using identities of diagnostic ions) in PC 16:1_18:1. By comparison, in PC 16:0_18:1, the relative amount of C18:1 (n-7) was only approximately 50%. Due to the high structure-resolving power of our method, we can even acquire the isomer composition of C16:1 or C18:1 in an sn-specific approach. The different sn-position of C16:1 or C18:1 fatty acyl does not seem to affect its C
C location isomers (n-7, n-9, and n-10) for phospholipids containing a C18:1 and C16:1 fatty acyl. Notably, the proportion of C18:1 (n-7) is greater than 80% (calculated using identities of diagnostic ions) in PC 16:1_18:1. By comparison, in PC 16:0_18:1, the relative amount of C18:1 (n-7) was only approximately 50%. Due to the high structure-resolving power of our method, we can even acquire the isomer composition of C16:1 or C18:1 in an sn-specific approach. The different sn-position of C16:1 or C18:1 fatty acyl does not seem to affect its C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C isomer compositions (Fig. 5b). A significant advantage of isomer analysis is its low coefficient of variation (CV). This may stem from the similar ionization and fragmentation efficiencies of the isomers and the fact that fluctuations in these efficiencies between individual cells are cancelled out through ratio calculation. This implies that more reproducible quantitative data for single-cell analysis can be obtained via ratio-based normalization, thereby achieving better quality control and laying a solid foundation for subsequent applied research.
C isomer compositions (Fig. 5b). A significant advantage of isomer analysis is its low coefficient of variation (CV). This may stem from the similar ionization and fragmentation efficiencies of the isomers and the fact that fluctuations in these efficiencies between individual cells are cancelled out through ratio calculation. This implies that more reproducible quantitative data for single-cell analysis can be obtained via ratio-based normalization, thereby achieving better quality control and laying a solid foundation for subsequent applied research.
Fig. 5c illustrates the sn-position isomer composition of representative phospholipids with fatty acyls identified. For example, PC 34:2 is a mixture of two pairs of sn-isomers: PC 16:1/18:1, PC 18:1/16:1, PC 16:0/18:2, and PC 18:2/16:0. These results are consistent with existing studies and confirm the robustness of the method for qualitative and quantitative analysis at the single-cell lipidome level.31
Distinguishing DOX-resistant and DOX-sensitive CML K562 cells
DOX is a widely utilized, broad-spectrum anti-tumor agent in leukemia therapy. Despite initial treatment efficacy, some patients can develop acquired DOX resistance. Accurate differentiation between DOX-resistant and -sensitive K562 cells in heterogeneous populations is crucial. Although there is sufficient evidence that DOX resistance is closely related to membrane proteins,42,43 insufficient research has been conducted on the cellular lipidome.44 In this section, we aim to differentiate DOX-resistant and DOX-sensitive CML K562 cells by analyzing their lipidomes at the single-cell level. DOX-resistant K562 cells were developed through prolonged exposure to 1 µg mL−1 DOX over 20 passages, while wild-type K562 cells, which were DOX-sensitive, served as controls.A targeted lipidomics analysis focused on 24 high-abundance and 20 low-abundance lipids. A total of 30 PCs, 14 PEs, and 8 PSs were detected at the sum composition level (Table S11), along with 30 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers and 23 sn-position isomers (Tables S12 and S13). The lipid C
C location isomers and 23 sn-position isomers (Tables S12 and S13). The lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location and sn-position isomers were expressed as isomer ratios to improve data robustness and quantitative accuracy (Table S14).41 The resulting dataset, i.e., a total of 96 dimensions including 51 lipid sum compositions (excluding PC 34:1), 22 C
C location and sn-position isomers were expressed as isomer ratios to improve data robustness and quantitative accuracy (Table S14).41 The resulting dataset, i.e., a total of 96 dimensions including 51 lipid sum compositions (excluding PC 34:1), 22 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations, 13 sn-positions, and 10 C
C locations, 13 sn-positions, and 10 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/sn-positions, was used for comparative analysis.
C/sn-positions, was used for comparative analysis.
The t-distributed stochastic neighbor embedding (t-SNE) algorithm and k-means method were employed to cluster DOX-resistant and -sensitive cells. To assess the effectiveness of different lipid features for distinguishing cell types, clustering analyses were conducted using various factors, including lipid sum composition, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location, sn-position, and their combinations (Fig. 6a and S22). Lipid composition alone failed to accurately discriminate the two cell types, with approximately 50% overlap between the two clusters. By comparison, it was found that the C
C location, sn-position, and their combinations (Fig. 6a and S22). Lipid composition alone failed to accurately discriminate the two cell types, with approximately 50% overlap between the two clusters. By comparison, it was found that the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location is a highly discriminating feature, highlighting the importance of single-cell structural lipidomics in biological applications. With multiple lipidomic features available, it can be expected that their combination could further improve the accuracy of cell classification.
C location is a highly discriminating feature, highlighting the importance of single-cell structural lipidomics in biological applications. With multiple lipidomic features available, it can be expected that their combination could further improve the accuracy of cell classification.
Cluster 1 was tentatively identified as DOX-resistant cells, with Cluster 2 identified as DOX-sensitive cells. Across all analyses shown in Fig. 5a, three cells are consistently misclassified (Fig. 5a and S22a–c). Specifically, cell #8(C1), a DOX-resistant cell, was incorrectly classified as DOX-sensitive, and cells #33(C2) and #38(C3), both DOX-sensitive, were misclassified as DOX-resistant. This resulted in an overall classification accuracy of 94.9%. The discrepancy of C2 and C3 cells might be attributed to the presence of drug-resistant subpopulations in the DOX-sensitive cells, aligning with the known existence of a small proportion of DOX-resistant cells within the otherwise sensitive populations.45
Lipidomic analysis of the remaining cell populations, excluding misclassified cells, revealed significant differences in lipid classes, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations, sn-positions, and C
C locations, sn-positions, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations at specific sn-positions (Fig. 6b). Volcano plot analysis identified 20 lipid features that are most significant to discriminate DOX-resistant and -sensitive cells (p < 10−5), including four molecular lipid species, seven lipid C
C locations at specific sn-positions (Fig. 6b). Volcano plot analysis identified 20 lipid features that are most significant to discriminate DOX-resistant and -sensitive cells (p < 10−5), including four molecular lipid species, seven lipid C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers, five sn-position isomers, and four C
C location isomers, five sn-position isomers, and four C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location isomers with identified fatty acyl sn-positions ( Fig. S23 and S24). For example, the n-10/n-7 isomer ratio of PC 16:0_16:1 was reduced mainly in DOX-resistant K562 cells (Fig. 6c), and the PE 18:1/18:1 to PE 18:2/18:0 ratio for PE 36:2 was also lower in DOX-resistant cells (Fig. 6c). Conversely, the C18:1 n-9/n-7 ratio for PC 16:1/18:1 was much higher in DOX-resistant cells (Fig. 6c). While the biological implications of these lipidome changes require further investigation, this study demonstrates that our single-cell lipidomics method is a powerful tool for analyzing cellular lipid heterogeneity and evaluating the drug resistance of cancer cells.
C location isomers with identified fatty acyl sn-positions ( Fig. S23 and S24). For example, the n-10/n-7 isomer ratio of PC 16:0_16:1 was reduced mainly in DOX-resistant K562 cells (Fig. 6c), and the PE 18:1/18:1 to PE 18:2/18:0 ratio for PE 36:2 was also lower in DOX-resistant cells (Fig. 6c). Conversely, the C18:1 n-9/n-7 ratio for PC 16:1/18:1 was much higher in DOX-resistant cells (Fig. 6c). While the biological implications of these lipidome changes require further investigation, this study demonstrates that our single-cell lipidomics method is a powerful tool for analyzing cellular lipid heterogeneity and evaluating the drug resistance of cancer cells.
Variations in the cellular lipidome at multiple structural levels during ferroptosis
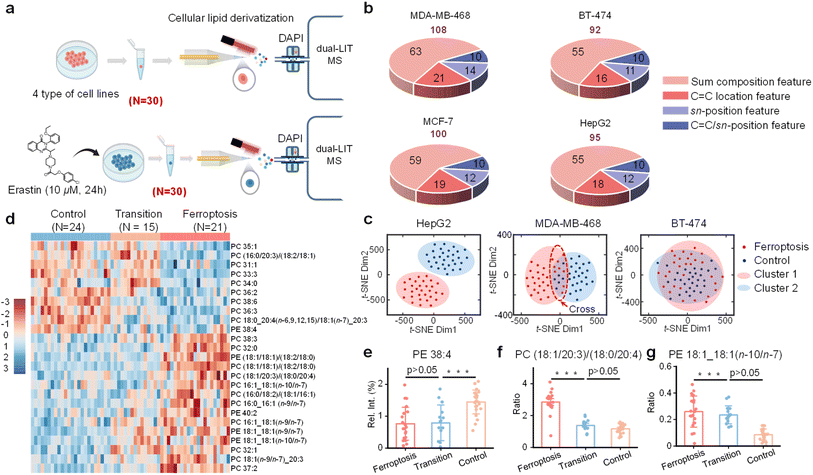
Ferroptosis is emerging as a critical factor in cancer, offering a potential therapeutic target for inducing cancer cell death and insights into the mechanisms of tumor suppression and chemoresistance.46–49 Ferroptosis can be triggered by inhibiting glutathione peroxidase 4 (GPX4) with small-molecule compounds, leading to lipid peroxidation through the accumulation of reactive oxygen species.50,51 While the link between lipid peroxidation and ferroptosis is well established, the connection between ferroptosis and lipid remodeling remains to be explored at the structural lipidomics level, e.g., at C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location and sn-position levels. For this purpose, we induced ferroptosis in multiple cancer cell lines (MDA-MB-468, MCF-7, BT-474, and HepG2) using erastin, a highly potent ferroptosis inducer (Fig. 7a). Lipid species identified from HepG2 are shown in Tables S15–S17. The single-cell lipidomics data are subjected to data conversion, previously used for the analysis of K562 cells, for comparative analysis (Fig. 7b).
C location and sn-position levels. For this purpose, we induced ferroptosis in multiple cancer cell lines (MDA-MB-468, MCF-7, BT-474, and HepG2) using erastin, a highly potent ferroptosis inducer (Fig. 7a). Lipid species identified from HepG2 are shown in Tables S15–S17. The single-cell lipidomics data are subjected to data conversion, previously used for the analysis of K562 cells, for comparative analysis (Fig. 7b).
Among all four cell lines after ferroptosis induction, HepG2 cells exhibited the most pronounced ferroptotic response, as evidenced by the significant alterations in single-cell lipidome profiles. t-SNE classification of the single-cell lipid data for the HepG2 cell population revealed distinct clustering patterns before and after ferroptosis (Fig. 7c). By contrast, BT-474 and MCF-7 cells were relatively insensitive to ferroptosis (Fig. 7c and S26). MDA-MB-468 cells were sensitive to ferroptosis, but the two cell clusters overlapped (Fig. 7c). These findings aligned with those of previous studies (Fig. S25).52 Hierarchical clustering of MDA-MB-468, performed without a pre-set number of classes, identified three cell groups: control, ferroptosis, and a transition group in the overlapped regions (Fig. S27). The transition group included nine cells from the ferroptosis group and six from the control group. Re-clustering confirmed the transition group to be intermediate between the control and ferroptosis groups (Fig. S28).
Upon further re-hierarchical clustering (Fig. 7d), it was observed that PE 38:4 levels were significantly lower in the ferroptosis group compared to the control, but similar to those in the transition group (Fig. 7e). This is consistent with the fact that PE 38:4 (i.e., PE 18:0_20:4) is depleted by lipid peroxidation during ferroptosis.50 The significantly higher ratio of PC (18:1/18:3)/(16:0/20:4) in the ferroptosis group is also consistent with C20:4 depletion (Fig. 7f). We also noted lipid changes at the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C level. For instance, in PE 18:1_18:1, the n-10/n-7 ratio was higher in the ferroptosis and transition groups than in the control group, possibly suggesting a relatively higher activity of fatty acid desaturase 2 (FADS2) (Fig. 7g). Elevated levels of C18:1 n-9/n-7 ratios were also observed in multiple lipid species, suggesting an altered interplay between stearoyl-CoA desaturase 1 (SCD1) and β oxidation.53,54
C level. For instance, in PE 18:1_18:1, the n-10/n-7 ratio was higher in the ferroptosis and transition groups than in the control group, possibly suggesting a relatively higher activity of fatty acid desaturase 2 (FADS2) (Fig. 7g). Elevated levels of C18:1 n-9/n-7 ratios were also observed in multiple lipid species, suggesting an altered interplay between stearoyl-CoA desaturase 1 (SCD1) and β oxidation.53,54
The transition group cells, which accounted for 25% of the total cell population, might represent a state of ferroptosis resistance. This ratio was consistent with those reported in previous studies on ferroptosis sensitivity in various cell populations.52 The distinct ‘Transition’ state may represent the asynchronous onset of ferroptosis, where cells initiate lipid peroxidation at different times. This is reflected in the heterogeneous lipidome profile (Fig. 7d–g), a mixture of cells depleting susceptible lipids (such as PE 38:4) alongside those that have not. Furthermore, this state could involve alternative lipid remodeling pathways. This transitional population, resisting full ferroptotic commitment, is analogous to partial drug resistance, linking our findings to broader cellular stress responses.
Conclusion
Lipidomics analyses of bulk samples have achieved significant developments, identifying over 1000 lipid species at the level of sum compositions, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C location, and sn-position.55,56 However, single-cell lipidomics faces unique challenges. Limited sample content from single mammalian cells restricts the number of feasible MS/MS analyses, and low ion utilization poses significant obstacles to comprehensive lipid structural characterization and accurate quantitative analysis. To address these limitations, we developed a high-coverage single-cell structural lipidomics method utilizing dual-LIT MS coupled with single-cell electrospray technology and online photochemical derivatization. The dual-LIT MS system, with its efficient ion storage capabilities, maximizes ion utilization and offers versatile MSn capabilities crucial for comprehensive lipid structure characterization.
C location, and sn-position.55,56 However, single-cell lipidomics faces unique challenges. Limited sample content from single mammalian cells restricts the number of feasible MS/MS analyses, and low ion utilization poses significant obstacles to comprehensive lipid structural characterization and accurate quantitative analysis. To address these limitations, we developed a high-coverage single-cell structural lipidomics method utilizing dual-LIT MS coupled with single-cell electrospray technology and online photochemical derivatization. The dual-LIT MS system, with its efficient ion storage capabilities, maximizes ion utilization and offers versatile MSn capabilities crucial for comprehensive lipid structure characterization.
Overall, this method represents a significant advancement and innovation in several aspects. (1) Enhanced ion utilization and MSn capabilities: the current study focuses on significantly improving ion utilization efficiency and expanding the capabilities of MSn analysis. We optimized the dual-LIT MS system, particularly the vacuum regulation system, to achieve efficient ion storage for over 30 s with minimal ion loss. This enhancement allows for multiple rounds of MSn analysis (n = 2–4) for each lipid species, enabling comprehensive structural characterization at different levels. We also implemented a PB-MS3/MS4 analysis strategy for comprehensive phospholipid characterization. This approach enables the determination of sn-specific fatty acyls and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C locations within them, providing the most in-depth analysis of phospholipids within single cells to date. (2) Targeted lipidomics strategy for high coverage: we developed a targeted lipidomics strategy to overcome the challenges of ion interference and maximize the coverage of lipid species within single cells. This strategy involves the separate analysis of high- and low-abundance lipid ions, using the MMPT mode for low-abundance ions and separate ion injection for high-abundance ions. This approach significantly improves the dynamic range of detectable lipids and enables the identification of 100+ distinct lipid molecules from a single cell. (3) Application to cell drug resistance and ferroptosis: we successfully applied the developed method to distinguish between DOX-resistant and DOX-sensitive K562 cells with high accuracy and to investigate ferroptosis in multiple cancer cell lines. These applications demonstrate the significant potential of this novel single-cell structural lipidomics method for advancing research on cell drug resistance and programmed cell death. In summary, we anticipate that this technology will have broad prospects for advancing deep single-cell lipidomic analysis and its applications.
C locations within them, providing the most in-depth analysis of phospholipids within single cells to date. (2) Targeted lipidomics strategy for high coverage: we developed a targeted lipidomics strategy to overcome the challenges of ion interference and maximize the coverage of lipid species within single cells. This strategy involves the separate analysis of high- and low-abundance lipid ions, using the MMPT mode for low-abundance ions and separate ion injection for high-abundance ions. This approach significantly improves the dynamic range of detectable lipids and enables the identification of 100+ distinct lipid molecules from a single cell. (3) Application to cell drug resistance and ferroptosis: we successfully applied the developed method to distinguish between DOX-resistant and DOX-sensitive K562 cells with high accuracy and to investigate ferroptosis in multiple cancer cell lines. These applications demonstrate the significant potential of this novel single-cell structural lipidomics method for advancing research on cell drug resistance and programmed cell death. In summary, we anticipate that this technology will have broad prospects for advancing deep single-cell lipidomic analysis and its applications.
Conflicts of interest
The authors declare the following competing financial interest(s): Zheng Ouyang is the founder of PURSPEC Technologies Inc., which develops miniature mass spectrometry systems.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: the single-cell experiment procedure, modes of ion manipulation via the dual-LIT-MS, single cell lipidome analysis via PB reactions, and lists of lipids identified. See DOI: https://doi.org/10.1039/d5sc08665e.Acknowledgements
This work was supported by the Beijing Natural Science Foundation (Grant: 2232007), the National Key R&D Program of China (Grant: 2022YFC3401900), and the National Natural Science Foundation of China (Grants: 21934003 and 22574092).References
- S. J. Altschuler and L. F. Wu, Cell, 2010, 141, 559–563 Search PubMed.
- D. G. Spiller, C. D. Wood, D. A. Rand and M. R. White, Nature, 2010, 465, 736–745 Search PubMed.
- F. A. Rosenberger, M. Thielert and M. Mann, Nat. Methods, 2023, 20, 320–323 Search PubMed.
- T. Wälchli, M. Ghobrial, M. Schwab, S. Takada, H. Zhong, S. Suntharalingham, S. Vetiska, D. R. Gonzalez, R. Wu, H. Rehrauer, A. Dinesh, K. Yu, E. L. Y. Chen, J. Bisschop, F. Farnhammer, A. Mansur, J. Kalucka, I. Tirosh, L. Regli, K. Schaller, K. Frei, T. Ketela, M. Bernstein, P. Kongkham, P. Carmeliet, T. Valiante, P. B. Dirks, M. L. Suva, G. Zadeh, V. Tabar, R. Schlapbach, H. W. Jackson, K. De Bock, J. E. Fish, P. P. Monnier, G. D. Bader and I. Radovanovic, Nature, 2024, 632, 603–613 Search PubMed.
- A. J. Ibanez, S. R. Fagerer, A. M. Schmidt, P. L. Urban, K. Jefimovs, P. Geiger, R. Dechant, M. Heinemann and R. Zenobi, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 8790–8794 Search PubMed.
- R. Zenobi, Science, 2013, 342, 1243259 Search PubMed.
- C. Seydel, Nat. Methods, 2021, 18, 1452–1456 Search PubMed.
- H. Zhu, Q. Li, T. Liao, X. Yin, Q. Chen, Z. Wang, M. Dai, L. Yi, S. Ge, C. Miao, W. Zeng, L. Qu, Z. Ju, G. Huang, C. Cang and W. Xiong, Nat. Methods, 2021, 18, 788–798 Search PubMed.
- A. Ali, S. Davidson, E. Fraenkel, I. Gilmore, T. Hankemeier, J. A. Kirwan, A. N. Lane, I. Lanekoff, M. Larion, L. I. McCall, M. Murphy, J. V. Sweedler and C. Zhu, Metabolomics, 2022, 18, 77 Search PubMed.
- Q. Zhang, Y. Tamura, M. Roy, Y. Adachi, M. Iijima and H. Sesaki, Cell. Mol. Life Sci., 2014, 71, 3767–3778 Search PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 Search PubMed.
- E. M. Storck, C. Özbalci and U. S. Eggert, Annu. Rev. Biochem., 2018, 87, 839–869 Search PubMed.
- Q. Liu, W. Ge, T. Wang, J. Lan, S. Martinez-Jarquin, C. Wolfrum, M. Stoffel and R. Zenobi, Angew. Chem., Int. Ed., 2021, 60, 24534–24542 Search PubMed.
- T. Xu, H. Li, P. Dou, Y. Luo, S. Pu, H. Mu, Z. Zhang, D. Feng, X. Hu, T. Wang, G. Tan, C. Chen, H. Li, X. Shi, C. Hu and G. Xu, Adv. Sci., 2024, 11, e2306659 Search PubMed.
- X. Zhao, J. Liang, Z. Chen, R. Jian, Y. Qian, Y. Wang, Z. Guo, W. Zhang, Y. Zhang, H. Yin and Y. Xia, Angew. Chem., Int. Ed., 2023, 62, e202215556 Search PubMed.
- J. P. Menzel, R. S. E. Young, A. H. Benfield, J. S. Scott, P. Wongsomboon, L. Cudlman, J. Cvacka, L. M. Butler, S. T. Henriques, B. L. J. Poad and S. J. Blanksby, Nat. Commun., 2023, 14, 3940 Search PubMed.
- T. J. Gu, P. K. Liu, Y. W. Wang, M. T. Flowers, S. Xu, Y. Liu, D. B. Davis and L. Li, Nat. Chem., 2024, 16, 762–770 Search PubMed.
- G. Feng, M. Gao, L. Wang, J. Chen, M. Hou, Q. Wan, Y. Lin, G. Xu, X. Qi and S. Chen, Nat. Commun., 2022, 13, 2652 Search PubMed.
- S. Tang, H. Wang, H. Zhang, M. Zhang, J. Xu, C. Yang, X. Chen and X. Guo, J. Am. Chem. Soc., 2024, 146, 29503–29512 Search PubMed.
- X. Ma, L. Chong, R. Tian, R. Shi, T. Y. Hu, Z. Ouyang and Y. Xia, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2573–2578 Search PubMed.
- P. Morigny, J. Boucher, P. Arner and D. Langin, Nat. Rev. Endocrinol., 2021, 17, 276–295 Search PubMed.
- S. Cheng, D. Zhang, J. Feng, Q. Hu, A. Tan, Z. Xie, Q. Chen, H. Huang, Y. Wei, Z. Ouyang and X. Ma, Research, 2023, 6, 0087 Search PubMed.
- A. S. Mutlu, J. Duffy and M. C. Wang, Dev. Cell, 2021, 56, 1394–1407 Search PubMed.
- S. A. Lim, W. Su, N. M. Chapman and H. Chi, Nat. Chem. Biol., 2022, 18, 470–481 Search PubMed.
- S. Oh, C. Lee, W. Yang, A. Li, A. Mukherjee, M. Basan, C. Ran, W. Yin, C. J. Tabin, D. Fu, X. S. Xie and M. W. Kirschner, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2117938119 Search PubMed.
- S. N. Thomas, D. French, P. J. Jannetto, B. A. Rappold and W. A. Clarke, Nat. Rev. Methods Primers, 2022, 2, 96 Search PubMed.
- S. Qin, Y. Zhang, M. Shi, D. Miao, J. Lu, L. Wen and Y. Bai, Nat. Commun., 2024, 15, 350 Search PubMed.
- H. Zhang, Y. Liu, L. Fields, X. Shi, P. Huang, H. Lu, A. J. Schneider, X. Tang, L. Puglielli, N. V. Welham and L. Li, Nat. Commun., 2023, 14, 5185 Search PubMed.
- Z. Li, S. Cheng, Q. Lin, W. Cao, J. Yang, M. Zhang, A. Shen, W. Zhang, Y. Xia, X. Ma and Z. Ouyang, Nat. Commun., 2021, 12, 2869 Search PubMed.
- M. R. L. Paine, B. L. J. Poad, G. B. Eijkel, D. L. Marshall, S. J. Blanksby, R. M. A. Heeren and S. R. Ellis, Angew. Chem., Int. Ed., 2018, 57, 10530–10534 Search PubMed.
- W. Cao, S. Cheng, J. Yang, J. Feng, W. Zhang, Z. Li, Q. Chen, Y. Xia, Z. Ouyang and X. Ma, Nat. Commun., 2020, 11, 375 Search PubMed.
- X. Liu, X. Wang, J. Bu, X. Zhou and Z. Ouyang, Anal. Chem., 2019, 91, 1391–1398 Search PubMed.
- X. Guo, W. Cao, X. Fan, Z. Guo, D. Zhang, H. Zhang, X. Ma, J. Dong, Y. Wang, W. Zhang and Z. Ouyang, Angew. Chem., Int. Ed., 2022, e202214804, DOI:10.1002/anie.202214804.
- X. C. Zhang, Z. W. Wei, X. Y. Gong, X. Y. Si, Y. Y. Zhao, C. D. Yang, S. C. Zhang and X. R. Zhang, Sci. Rep., 2016, 6, 24730 Search PubMed.
- Y. Cong, K. Motamedchaboki, S. A. Misal, Y. Liang, A. J. Guise, T. Truong, R. Huguet, E. D. Plowey, Y. Zhu, D. Lopez-Ferrer and R. T. Kelly, Chem. Sci., 2021, 12, 1001–1006 Search PubMed.
- P. Mandal, S. Das, D. De Munshi, T. Dutta and M. Mukherjee, Int. J. Mass Spectrom., 2014, 364, 16–20 Search PubMed.
- N. Li, X. Zhou and Z. Ouyang, Int. J. Mass Spectrom., 2021, 462, 116523 Search PubMed.
- M. H. Soni and R. G. Cooks, Anal. Chem., 1994, 66, 2488–2496 Search PubMed.
- Z. Xu, T. Jiang, Q. Xu, Y. Zhai, D. Li and W. Xu, Anal. Chem., 2019, 91, 13838–13846 Search PubMed.
- Y. Zhu, W. Wang and Z. Yang, Anal. Chem., 2020, 92, 11380–11387 Search PubMed.
- S. Cheng, C. Cao, Y. Qian, H. Yao, X. Gong, X. Dai, Z. Ouyang and X. Ma, Chem. Sci., 2024, 15, 6314–6320 Search PubMed.
- H. Hamada and T. Tsuruo, Cancer Res., 1988, 48, 4926–4932 Search PubMed.
- M. Szwed, K. D. Kania and Z. Jozwiak, Cell. Oncol., 2014, 37, 421–428 Search PubMed.
- T. C. Karagiannis, M. Wall, K. Ververis, E. Pitsillou, S. M. Tortorella, P. A. Wood, H. Rafehi, I. Khurana, S. S. Maxwell, A. Hung, J. Vongsvivut and A. El-Osta, Cell. Mol. Life Sci., 2023, 80, 248 Search PubMed.
- S. Zhang, X. Liu, T. Bawa-Khalfe, L. S. Lu, Y. L. Lyu, L. F. Liu and E. T. Yeh, Nat. Med., 2012, 18, 1639–1642 Search PubMed.
- J. D. Scott, K. M. Lemberg, M. R. Lamprecht, R. Skouta, E. M. Zaitsev, C. E. Gleason, D. N. Patel, A. J. Bauer, A. M. Cantley, W. S. Yang, B. Morrison and B. R. Stockwell, Cell, 2012, 149, 1060–1072 Search PubMed.
- J. P. Friedmann Angeli, D. V. Krysko and M. Conrad, Nat. Rev. Cancer, 2019, 19, 405–414 Search PubMed.
- X. Jiang, B. R. Stockwell and M. Conrad, Nat. Rev. Mol. Cell Biol., 2021, 22, 266–282 Search PubMed.
- S. J. Dixon and J. A. Olzmann, Nat. Rev. Mol. Cell Biol., 2024, 25, 424–442 Search PubMed.
- S. Doll, B. Proneth, Y. Y. Tyurina, E. Panzilius, S. Kobayashi, I. Ingold, M. Irmler, J. Beckers, M. Aichler, A. Walch, H. Prokisch, D. Trumbach, G. Mao, F. Qu, H. Bayir, J. Fullekrug, C. H. Scheel, W. Wurst, J. A. Schick, V. E. Kagan, J. P. Angeli and M. Conrad, Nat. Chem. Biol., 2017, 13, 91–98 Search PubMed.
- D. Tang, X. Chen, R. Kang and G. Kroemer, Cell Res., 2021, 31, 107–125 Search PubMed.
- B. Seashore-Ludlow, M. G. Rees, J. H. Cheah, M. Cokol, E. V. Price, M. E. Coletti, V. Jones, N. E. Bodycombe, C. K. Soule, J. Gould, B. Alexander, A. Li, P. Montgomery, M. J. Wawer, N. Kuru, J. D. Kotz, C. S. Hon, B. Munoz, T. Liefeld, V. Dancik, J. A. Bittker, M. Palmer, J. E. Bradner, A. F. Shamji, P. A. Clemons and S. L. Schreiber, Cancer Discovery, 2015, 5, 1210–1223 Search PubMed.
- B. Koletzko, E. Reischl, C. Tanjung, I. Gonzalez-Casanova, U. Ramakrishnan, S. Meldrum, K. Simmer, J. Heinrich and H. Demmelmair, Annu. Rev. Nutr., 2019, 39, 21–44 Search PubMed.
- R. S. E. Young, A. P. Bowman, E. D. Williams, K. D. Tousignant, C. L. Bidgood, V. R. Narreddula, R. Gupta, D. L. Marshall, B. L. J. Poad, C. C. Nelson, S. R. Ellis, R. M. A. Heeren, M. C. Sadowski and S. J. Blanksby, Cell Rep., 2021, 34, 108738 Search PubMed.
- T. Xia, F. Zhou, D. Zhang, X. Jin, H. Shi, H. Yin, Y. Gong and Y. Xia, Nat. Commun., 2023, 14, 4263 Search PubMed.
- J. A. Michael, R. S. E. Young, R. Balez, L. J. Jekimovs, D. L. Marshall, B. L. J. Poad, T. W. Mitchell, S. J. Blanksby, C. S. Ejsing and S. R. Ellis, Angew. Chem., Int. Ed., 2024, e202316793 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |