Open Access Article
Open Access ArticleGenetically encoded green-light-responsive photocaged lysine for sequential control of protein function
Manjia
Li†
a,
Minghao
Lu†
a,
Lijun
Wang
a,
Yuqing
Zhang
a,
Long
Yan
a,
Shushu
Wang
a and
Tao
Peng
 *ab
*ab
aSchool of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, China. E-mail: tpeng@pku.edu.cn
bInstitute of Chemical Biology, Shenzhen Bay Laboratory, Shenzhen 518132, China
First published on 25th November 2025
Abstract
Site-specific incorporation of photo-responsive unnatural amino acids (UAAs) into proteins via genetic code expansion offers a powerful approach to control and study protein function in biological systems. However, existing UAAs are all sensitive to UV or near-UV light, and no visible-light-responsive UAAs have been reported, limiting our ability to regulate multiple proteins simultaneously. Here, we present the genetic encoding of a green-light-activatable lysine derivative, SCouK, for sequential photocontrol of protein activities in live cells. SCouK, containing a photolabile thiocoumarin moiety at the Nε-amino group of lysine, can be genetically encoded into proteins in bacterial and mammalian cells. We show that site-specifically incorporated SCouK can be photoactivated across a broad wavelength range, from UV to green light, to restore the functions of EGFP and luciferase. Notably, SCouK is highly efficiently photodecaged by green light centered at 520 nm within 30 seconds, marking it as the first visible-light-responsive lysine derivative with the longest single-photon activation wavelength among genetically encoded photolabile UAAs. Additionally, we showcase the general capability of SCouK for the optical control of different kinases and temporal control and interrogation of the cGAS-STING pathway in live cells. Moreover, by combing SCouK with a UV-light-activatable tyrosine derivative, we achieve, for the first time, sequential photoactivation of two distinct UAA-modified proteins within a single live-cell sample. Overall, the unique features of SCouK, including site-specific incorporation, green-light-responsiveness, orthogonal activation wavelengths, high decaging efficiency, and general applicability, demonstrate its great potential to non-invasively and precisely manipulate proteins in complex living systems for functional studies and therapeutic applications.
Introduction
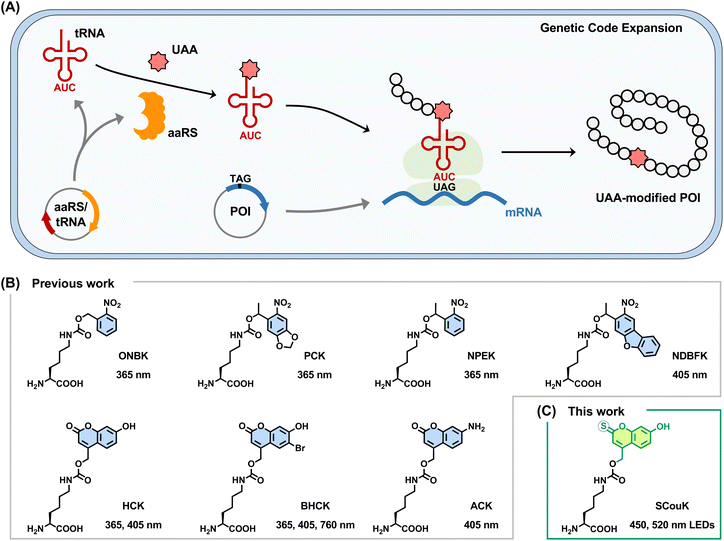
Optical tools, relying on the use of light, have emerged as a powerful way to study and manipulate biological processes in living systems, owing to many unique advantages including non-invasiveness, homogeneous distribution, easy control, bioorthogonality, and spatiotemporal precision.1–3 Specifically, photo-responsive small molecule inhibitors4,5 and photoswitchable ligands6,7 have been developed to probe protein function in complex cellular pathways. In addition, natural and engineered photo-responsive receptor proteins have been exploited to allow control of gene expression and cell behaviors in individual cells within a heterogeneous tissue.8A unique approach for the optical control of protein function and activity in live cells has been enabled by genetic code expansion.9–11 This technology utilizes orthogonal aminoacyl-tRNA synthetase (aaRS) and tRNA pairs to recognize unnatural amino acids (UAAs) and site-specifically incorporate them into proteins in response to nonsense codons, such as the amber stop codon (TAG), introduced into the corresponding genes (Fig. 1A).12–15 In this way, UAAs carrying unnatural chemical functionalities can be precisely positioned within proteins, providing powerful tools to probe and manipulate protein functions.16,17 In particular, a photocaged UAA bearing a photolabile protecting group is site-specifically introduced into the protein of interest via genetic code expansion at the key functional residue to temporarily block its activity. Upon light illumination, the photolabile group is selectively removed to restore the native functional residue, resulting in “switch-on” of the protein activity. Notable examples of such genetically encoded photocaged UAAs include derivatives of lysine,18–20 cysteine,21–23 tyrosine,24–26 serine,27 glutamic acid,28,29 aspartic acid,30 and histidine,31 which have been elegantly utilized to control various proteins, including kinases,32,33 DNA and RNA polymerases,34,35 tumor suppressor p53,19 virus capsid proteins,36 and antibodies,37–39 for manipulating cell signaling,32,33 enzymatic activity,34,35 protein localization,19 and protein–protein interaction.36–39
Among them, photocaged lysines are of particular interest given the essential roles of lysine in catalysis, binding, and recognition widely associated with many fundamental biological processes. In this regard, lysine derivatives containing o-nitrobenzyl-based photolabile groups (Fig. 1B), such as o-nitrobenzyl lysine (ONBK),18 4,5-methylenedioxy-o-nitrobenzyl lysine (PCK),19 α-methyl-o-nitrobenzyl lysine (NPEK),40 and nitrodibenzylfuranyl lysine (NDBFK),40 have been developed and utilized to optically control the functions of diverse proteins, including green fluorescent protein (GFP), luciferase, recombinase, and GTPase, in various contexts (Table S1 in SI). However, activation of these photocaged lysines generally requires high-energy ultraviolet (UV) light (e.g., 365 nm), which is limited in phototoxicity41 and poor tissue penetration.42 Moreover, o-nitrobenzyl-caged lysines could be decaged by reducing environments and enzymes (e.g., nitroreductase) in prokaryotic cells43,44 as well as in hypoxic mammalian cells,45 making them incompatible with some experiments. As alternatives, coumarin-caged lysines, such as hydroxycoumarin lysine (HCK),20 bromo-hydroxycoumarin lysine (BHCK),20 and aminocoumarin lysine (ACK),46 have been developed to allow photoactivation of proteins with near-UV light (e.g., 405 nm) or two-photon near-infrared (NIR) light (e.g., 760 nm) (Fig. 1B and Table S1 in SI). Despite these progresses, currently reported photocaged UAAs almost all require UV or near-UV light below 405 nm for activation, with only one exception (i.e., BHCK), which can be activated by two-photon NIR lasers.20 Photocaged UAAs that can be activated by visible light above 405 nm have not yet been reported. In comparison to UV light, which exhibits phototoxicity and limited tissue penetration, long-wavelength visible light is generally considered more biologically benign, offering reduced phototoxicity and deeper tissue penetration.47 More importantly, photocaged UAAs activatable by wavelengths orthogonal to those of UV and near-UV light could be particularly useful for sequential or orthogonal photocontrol of multiple protein targets in a single experiment within complex biological contexts. Due to the lack of such UAAs, sequential control of multiple UAA-modified proteins has not yet been achieved. Therefore, there is an urgent need to develop visible-light-activatable UAAs.
Herein, we present a thiocoumarin-derived lysine analogue, SCouK (Fig. 1C), that can be genetically encoded into proteins via genetic code expansion and efficiently uncaged by a broad range of wavelengths of light from UV and near-UV (e.g., 254 nm, 302 nm, 365 nm, and 390 nm) to blue (e.g., 450 nm) and green (e.g., 520 nm). Notably, SCouK is photoactivated rapidly (within 30 s) upon exposure to green light (centered at 520 nm, bandwidth ± 19 nm; 22 mW cm−2) that is orthogonal to existing photocaged UAAs, representing the first green-light-responsive lysine-derived UAA with great potential to reduce phototoxicity and enhance penetration. We demonstrate the broad utility of SCouK in optically controlling the functions and activities of various proteins, including GFP, luciferase, kinases, and nucleotidyltransferase, by genetically encoding SCouK at their essential lysine residues (Fig. 2A). More importantly, we apply SCouK to achieve the sequential two-wavelength photoactivation of two luciferases in live cells in a single experiment for the first time. We believe the unique photophysical properties of SCouK render it a valuable chemical tool for precisely manipulating and studying protein function in complex living systems.
Results and discussion
Design of a thiocoumarin-based photolabile lysine derivative SCouK
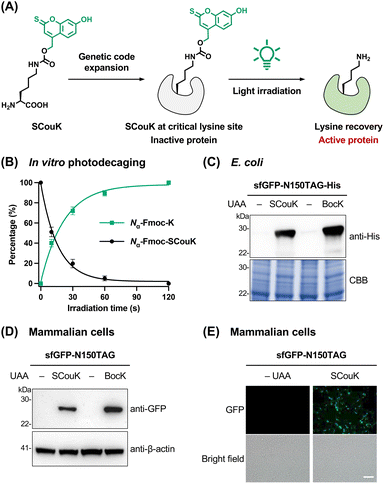
To develop new genetically encodable photocaged lysine, we sought to explore the thiocoumarin backbone due to its attractive features such as small size, synthetic accessibility, fast uncaging kinetics, low toxicity of the decaged byproduct, and red-shifted absorption compared to the carbonyl analogue.48 While the 7-diethylamino-2-thiocoumarin-4-ylmethyl moiety was used for photocaging small-molecule ligands,49–51 its 7-hydroxyl analogue has not been reported yet. We envisioned that the 7-hydroxy-2-thiocoumarin-4-ylmethyl group could be ideal for caging lysine due to its smaller size than the 7-diethylamino counterpart. In this regard, we designed a novel thiocoumarin-based lysine derivative, SCouK, by incorporating the 7-hydroxy-2-thiocoumarin-4-ylmethyl cage at the Nε-amino group of L-lysine through a carbamate linker (Fig. 2A). SCouK was synthesized and characterized as detailed in Scheme S1 in SI.SCouK exhibited a significantly red-shifted absorption maximum (λmax = 453 nm), and a broader and stronger absorption spectrum compared to its carbonyl analogue, i.e., HCK20 (Fig. S1A and B in SI). Meanwhile, the fluorescence of SCouK was markedly weaker than that of HCK (Fig. S1C and D in SI), minimizing potential interference with fluorescent probes such as GFP for cellular imaging studies. We expected that the thiocoumarin moiety, like the classic coumarin counterpart, absorbs photons to undergo photoheterolytic bond cleavage leading to the singlet ion pair,52 which further hydrolyzes to generate thiocoumarinylmethanol and the free lysine (Fig. S2 in SI). To validate the light-triggered removal of the 7-hydroxy-2-thiocoumarin-4-ylmethyl cage, we prepared Nα-Fmoc-protected SCouK (Nα-Fmoc-SCouK) that contains the same photocage as SCouK and additional aromatic groups for ease of detection by UV absorption (Fig. S3A in SI). The aqueous solution of Nα-Fmoc-SCouK in phosphate-buffered saline (PBS; 20 mM, pH 7.4) was irradiated for varying periods and analyzed by high-performance liquid chromatography-mass spectrometry (HPLC-MS). Interestingly, we found that irradiation of Nα-Fmoc-SCouK with a light-emitting diode (LED) bulb centered at 520 nm (bandwidth ± 19 nm; 22 mW cm−2) (Table S2 in SI) readily generated Nα-Fmoc-K and thiocoumarinylmethanol (Fig. S3B and C in SI). The yield of Nα-Fmoc-K was calculated to be about 40% after a 10 s light exposure and almost plateaued after irradiation for 1 min (Fig. 2B). The photodecaging half-life (t1/2), defined as the irradiation time required for 50% decaging, of Nα-Fmoc-SCouK in PBS (20 mM, pH 7.4) was determined to be 11.2 ± 1.6 s (mean ± SD, n = 4). In addition, Nα-Fmoc-SCouK exhibited pH-dependent photodecaging under 520 nm irradiation, showing lower efficiency at acidic pH and higher efficiency at basic pH (Fig. S4A in SI). This trend is consistent with the pH-dependent absorption of the 7-hydroxy-2-thiocoumarin moiety, which displays an apparent pKa of 7.0 (Fig. S4B and C in SI). Notably, efficient photodecaging was observed for Nα-Fmoc-SCouK at physiological pH 7.4, despite the limited yet apparent overlap between the absorption spectrum of SCouK and the emission spectrum of the 520 nm LED (Fig. S5A in SI). A weak absorption shoulder of SCouK extending from 500 to 550 nm is present and overlaps with the LED emission, which could contribute to its photoactivation. This phenomenon of limited overlap can also be explained by the recently well-established red-shifted photochemical reactivity relative to absorption,53–55 where the wavelength of maximal photochemical efficiency does not necessarily coincide with the absorption maximum but often shifts to longer wavelengths. Consistent with this, similar behaviors have been reported for other photocages, including o-nitrobenzyl derivatives53 and HCK, which shows minimal absorption at 405 nm (Fig. 1B in SI) but readily undergoes activation under 405 nm irradiation.20 Importantly, the emission spectrum of the 520 nm LED confirmed that it is indeed a green light source, with blue emission below 490 nm contributing about 6% and red emission above 570 nm contributing only 2% of the total output (Fig. S5B in SI), supporting that SCouK is green-light responsive. Overall, these results demonstrate that the 7-hydroxy-2-thiocoumarin-4-ylmethyl cage on the lysine side chain can be efficiently removed upon irradiation with green light centered at 520 nm.
Genetic encoding of SCouK into proteins in bacterial and mammalian cells
After validating the photodecaging of SCouK, we then explored the site-specific incorporation of SCouK into proteins via the genetic code expansion technology. The Methanosarcina mazei (Mm) pyrrolysyl-tRNA synthetase (MmPylRS) and MmPyl-tRNACUA pair has been widely used to genetically encode various lysine-derived UAAs in different organisms.56–59 Considering the structural similarities between SCouK and our previously reported aminocoumarin-derived UAAs,60,61 we focused on a series of active-site mutants of MmPylRS that were previously generated in our laboratory.62–70 These MmPylRS mutants mainly cover the Y306 and L309 sites and contain the Y384F mutation to improve the aminoacylation efficiency.71 To perform the screening, plasmids encoding MmPylRS mutants were individually co-transformed into E. coli together with the plasmid carrying enhanced GFP (EGFP) with an amber codon at the permissive Y39 residue (EGFP-Y39TAG) and a cognate MmPyl-tRNACUA (Fig. S6 and Table S3 in SI). The incorporation efficiency of SCouK was evaluated by the fluorescence intensity of full-length EGFP (Fig. S7A in SI). To our delight, the screening results demonstrated that SCouK can be efficiently incorporated into EGFP-Y39TAG by multiple mutants and that the Y306A/L309A/Y384F/I405R mutant, termed SCouKRS afterwards, displayed the highest efficiency to direct the incorporation of SCouK (Fig. S7B and C in SI).To further assess the site-specific incorporation of SCouK into proteins in E. coli, we chose superfolder GFP (sfGFP) with an amber codon at the permissive N150 site (sfGFP-N150TAG) as the model protein. For this, the plasmid carrying His-tagged sfGFP-N150TAG and MmPyl-tRNACUA was co-transformed with the SCouKRS-encoding plasmid (Fig. S6 and Table S3 in SI) into E. coli. Western blotting analysis confirmed that full-length sfGFP was expressed in the presence of SCouK (Fig. 2C) in a concentration-dependent manner (Fig. S8 in SI). The incorporation efficiency of SCouK by SCouKRS was comparable to that of Nε-Boc-L-lysine (BocK),72 which serves as an excellent substrate of wild-type MmPylRS and was incorporated by this synthetase in our comparison (Fig. 2C). We purified the sfGFP-N150SCouK protein from E. coli at a yield of 7.8 mg L−1, which is approximate to the yields of EGFP variants incorporated with BocK or aminocoumarin-derived UAAs.60 More importantly, MS analysis demonstrated that the purified sfGFP-N150SCouK protein has an observed mass of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 075.30 Da, in consistence with the calculated mass of 28
075.30 Da, in consistence with the calculated mass of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 075.28 Da (Fig. S9A–C in SI). Therefore, these data verify that SCouK is site-specifically incorporated into sfGFP in E. coli in high efficiency and fidelity. To further examine the photoinduced decaging of SCouK at the protein level in vitro, the purified sfGFP-N150SCouK protein was briefly exposed to green light irradiation (520 nm, bandwidth ± 19 nm; 22 mW cm−2) for 30 s and analyzed by MS. Notably, the observed mass of the resulting protein was shifted to 27
075.28 Da (Fig. S9A–C in SI). Therefore, these data verify that SCouK is site-specifically incorporated into sfGFP in E. coli in high efficiency and fidelity. To further examine the photoinduced decaging of SCouK at the protein level in vitro, the purified sfGFP-N150SCouK protein was briefly exposed to green light irradiation (520 nm, bandwidth ± 19 nm; 22 mW cm−2) for 30 s and analyzed by MS. Notably, the observed mass of the resulting protein was shifted to 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840.56 Da (Fig. S9D and E in SI), which is consistent with the theoretical mass of sfGFP-N150K (27
840.56 Da (Fig. S9D and E in SI), which is consistent with the theoretical mass of sfGFP-N150K (27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 841.39 Da). These results thus demonstrate the feasibility of photoinduced decaging of SCouK at the protein level.
841.39 Da). These results thus demonstrate the feasibility of photoinduced decaging of SCouK at the protein level.
We also examined the genetic encoding of SCouK in mammalian cells. Specifically, the plasmid encoding sfGFP-N150TAG was co-transfected with the plasmid carrying SCouKRS and its cognate MmPyl-tRNACUA (Fig. S10 and Table S3 in SI) into HEK293T cells to express the SCouK-containing sfGFP variant. Western blotting analysis showed that the full-length sfGFP protein was expressed in the presence of SCouK (Fig. 2D) in a concentration-dependent manner (Fig. S11A in SI). Consistent with the results in E. coli, the incorporation efficiency of SCouK by SCouKRS in mammalian cells was comparable to that of BocK, which was incorporated by wild-type MmPylRS (Fig. 2D). Fluorescence imaging analysis confirmed the SCouK-dependent expression of full-length sfGFP in HEK293T cells (Fig. 2E). Both western blotting and fluorescence imaging analyses showed that 0.2 mM of SCouK was sufficient to induce the optimal expression of sfGFP-N150SCouK in HEK293T cells (Fig. S11 in SI). Although coumarin derivatives are known to exhibit certain biological activities, no significant cell death or morphological changes were observed under these conditions. Consistently, cytotoxicity assays confirmed that both SCouK and its decaging product (generated in situ upon light irradiation) had no detectable effects on cell viability over 24 h (Fig. S12 in SI). Together, these results suggest that SCouK can be site-specifically encoded into proteins with high efficiency and biocompatibility in mammalian cells.
Optical control of EGFP fluorescence using SCouK
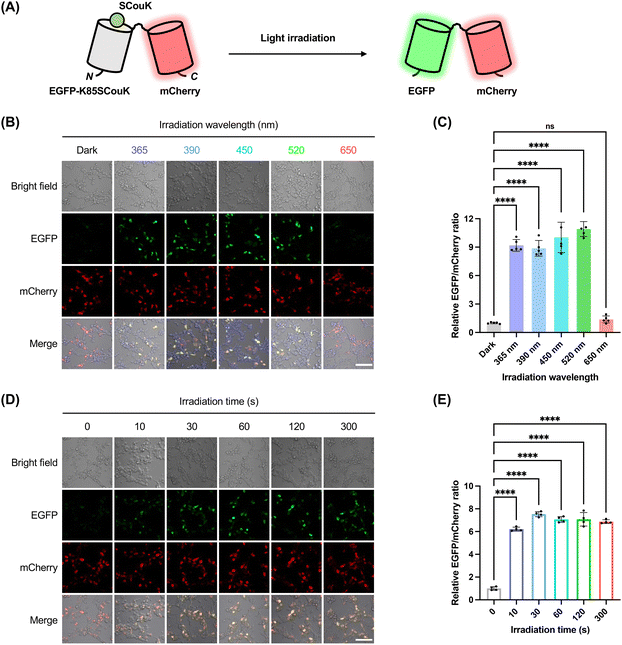
We next proceeded to investigate whether site-specifically incorporated SCouK could be used to optically control protein activity in live mammalian cells. As the first demonstration, we sought to generate a photo-activatable EGFP variant by replacing the key lysine residue of EGFP with SCouK. We and others have previously shown that the K85 residue is important for the maturation of EGFP chromophore.20,63,64 It was thus expected that substitution of K85 with SCouK could abolish EGFP fluorescence, whereas the photolysis of SCouK could restore the lysine residue to recover its fluorescence (Fig. 3A). In particular, we co-transfected HEK293T cells with two plasmids, i.e., the EGFP-K85TAG-mCherry-encoding plasmid and the plasmid carrying SCouKRS and MmPyl-tRNACUA (Fig. S10 and Table S3 in SI), to express the chimeric protein EGFP-K85SCouK-mCherry, in which an mCherry tag is fused to the C-terminus of EGFP-K85SCouK to serve as an internal standard. Fluorescence imaging analysis showed that the chimeric EGFP-K85SCouK-mCherry protein was well-expressed as indicated by the red fluorescence inside cells (Fig. 3B). As expected, no green fluorescence was observed before exposure of the cells to light, owing to the presence of SCouK at the essential K85 site of EGFP. This result confirms that the low intrinsic fluorescence of SCouK (Fig. S1C and D in SI) does not interfere with EGFP fluorescence during cellular imaging. By contrast, strong green fluorescence, overlapping with red fluorescence, was recorded in cells that were irradiated with a 520 nm green LED (bandwidth ± 19 nm; 22 mW cm−2) (Fig. 3B), attributable to photodecaging of SCouK to restore the lysine residue. Interestingly, comparably strong green fluorescence was also observed in cells after illumination with UV light at 365 nm or with LEDs centered at 390 nm or 450 nm (Fig. S13 and Table S2 in SI), but not with the red LED at 650 nm (Fig. 3B). Quantification of EGFP and mCherry fluorescence ratios (EGFP/mCherry) confirmed efficient decaging across wavelengths from 365 to 520 nm (i.e., UV to blue and green regions), with no detectable activation at 650 nm (Fig. 3C). Although the absorption spectrum of SCouK overlaps more strongly with the 450 nm LED than with the 520 nm LED (Fig. S5A and C in SI), the observed decaging efficiencies at these wavelengths were comparable, again highlighting the known mismatch between absorption spectra and wavelength-resolved photochemical reactivity.53–55 We next focused on the 520 nm LED to examine the effect of irradiation time on the decaging efficiency. For this, HEK293T cells expressing EGFP-K85SCouK-mCherry were exposed to the green light centered at 520 nm for different periods and monitored by fluorescence imaging. Activation of EGFP fluorescence was already significant after a brief irradiation (e.g., 10 s) and reached the plateau after a 30 s light exposure (Fig. 3D and E), suggesting the fast kinetics of photoinduced decaging of SCouK. These results also confirmed that mCherry-positive cells displayed no green fluorescence without irradiation, and mCherry-negative cells exhibited no EGFP fluorescence after irradiation, indicating the potential of SCouK for spatially resolved protein activation. To further demonstrate this, we transfected cells to express EGFP-K85SCouK-mCherry and applied localized light irradiation in a culture dish to activate EGFP. Only cells within the locally irradiated regions of the culture dish displayed strong green fluorescence, whereas cells in non-irradiated regions remained non-fluorescent (Fig. S14 in SI), confirming spatially restricted activation of SCouK-modified EGFP at the cellular level. Future integration with highly focused and localized optical illumination, such as region-of-interest (ROI) irradiation using laser scanning confocal microscopes, analogous to laser micro-irradiation,73 could open the possibility for selective activation of SCouK-modified proteins within defined regions at the subcellular level. Two-photon excitation, offering greater spatial precision and deeper tissue penetration,74 may further expand the applicability of SCouK for subcellular and deep-tissue activation. Given that HCK is non-responsive to two-photon excitation while its brominated analogue BHCK (Fig. 1B) is two-photon active,20 a brominated SCouK derivative could likewise enable two-photon control and should be investigated in future studies. Nonetheless, our current data demonstrate that SCouK enables optical control of EGFP fluorescence in live cells with green light centered at 520 nm.Optical control of luciferase activity using SCouK
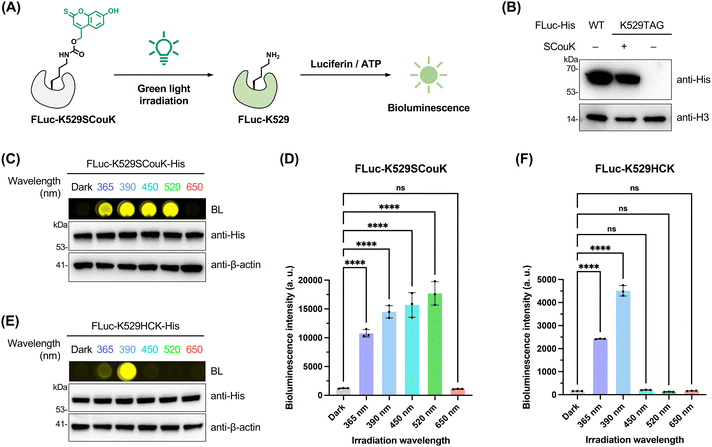
To investigate the capability of SCouK for the control of enzymatic activities in live cells, we firstly focused on firefly luciferase (FLuc). The K529 residue of FLuc stabilizes the intermediate formed during adenylation of the substrate through hydrogen-bonding with the phosphate group of ATP and is thus essential for the enzymatic activity of FLuc to produce bioluminescence (Fig. 4A).75 In this regard, HEK293T cells were co-transfected with a plasmid encoding FLuc-K529TAG and the plasmid encoding SCouKRS and MmPyl-tRNACUA (Fig. S10 and Table S3 in SI). Western blotting analysis confirmed the expression of full-length FLuc-K529SCouK protein in the presence of SCouK (Fig. 4B). The FLuc-K529SCouK-expressing cells were exposed to light sources at different wavelengths. Notably, no luminescence signal, that is, no FLuc activity, was detected in these cells before light irradiation (Fig. 4C), due to blocking of the catalytic activity of FLuc by SCouK at the K529 site. After irradiation with light centered at 365 nm, 390 nm, 450 nm, or 520 nm, strong bioluminescence signals were detected, whereas no activation occurred under the 650 nm light (Fig. 4C and D). Cells expressing FLuc-K529BocK showed no detectable luciferase activity before or after irradiation at any wavelength (Fig. S15A and B in SI). These results together confirm that incorporation and photodecaging of SCouK are required to restore the critical lysine residue and recover FLuc activity. Western blotting analysis of irradiated and nonirradiated cells confirmed comparable expression levels of full-length FLuc across different irradiation conditions without photo-induced protein degradation (Fig. 4C and S15A in SI).We compared the photodecaging behavior of SCouK with that of its carbonyl analogue HCK20 (Fig. 1B and Table S1 in SI), which differs from SCouK by only one atom and can be incorporated into FLuc-K529TAG at levels similar to SCouK and BocK (Fig. S15C in SI). As expected, cells expressing comparable levels of FLuc-K529HCK showed increased FLuc activity after exposure to light centered at 365 nm or 390 nm, but had no FLuc activity after irradiation with light centered at longer wavelengths, such as at 450 nm and 520 nm (Fig. 4E and F), indicating that HCK cannot be activated by blue or green light at these wavelengths. This difference highlights the essential role of the single sulfur atom in red-shifting the photodecaging wavelength.
Together, these data validate that site-specifically incorporated SCouK can be photolyzed by a broad range of wavelengths to activate protein function. Notably, SCouK is highly efficiently photoactivated using green light centered at 520 nm (bandwidth ± 19 nm; 22 mW cm−2), a wavelength that is orthogonal to those employed for currently available photocaged lysine, tyrosine, cysteine, and serine analogues.10
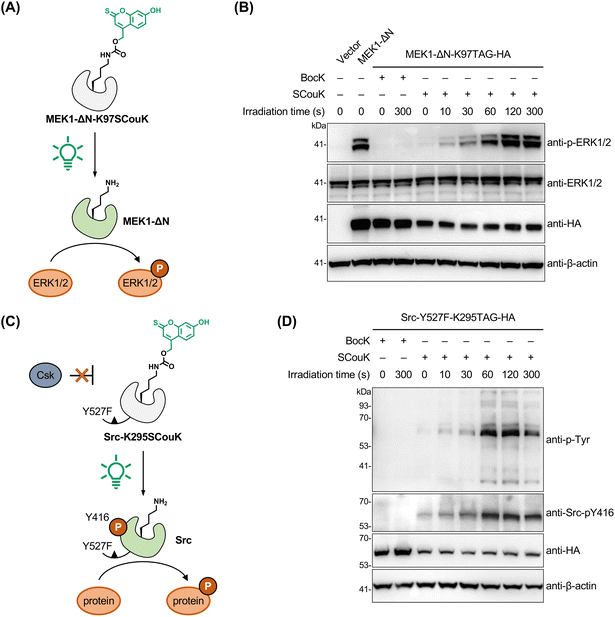
Optical control of kinase activities using SCouK
To demonstrate the general applicability of SCouK in optical activation of enzymes, we moved on to protein kinases, because they contain conserved catalytic lysine residues in the ATP-binding pockets critical for ATP binding and enzymatic activities. For this, we chose mitogen-activated protein kinase kinase 1 (MAPKK1, also known as MEK1) that is central to the MAPK signaling pathway and, upon activation, phosphorylates the downstream effectors such as extracellular signal-regulated kinase 1 and 2 (ERK1/2) for signaling transduction (Fig. 5A).76 To optically control MEK1 with SCouK and reduce the interference from endogenous upstream signaling, we utilized the constitutively active MEK1 variant bearing N-terminal deletion and S218D/S222D mutation (MEK1-ΔN)32 and replaced the catalytic lysine residue K97 with the TAG codon to generate the MEK1-ΔN-K97TAG variant. HEK293T cells were co-transfected with a plasmid encoding MEK1-ΔN-K97TAG and another plasmid encoding SCouKRS and MmPyl-tRNACUA (Fig. S10 and Table S3 in SI) in the presence of SCouK or BocK. The cells were briefly exposed to 520 nm green light (22 mW cm−2). Western blotting analysis confirmed the expression of HA-tagged MEK1-ΔN variants incorporated with SCouK or BocK (Fig. 5B). While no downstream ERK1/2 phosphorylation signals were observed in MEK1-ΔN-K97SCouK-expressing cells without light exposure, ERK1/2 phosphorylation levels were substantially increased upon a 10 s irradiation and plateaued after a 2 min irradiation with green light (Fig. 5B). By contrast, cells expressing the non-photoactivatable variant MEK1-ΔN-K97BocK had no ERK1/2 phosphorylation signals, no matter before or after light irradiation (Fig. 5B). These results thus demonstrate that SCouK in place of the catalytic K97 site allows optical control of MEK1 signaling in live cells.We also tested the applicability of SCouK in the optical control of another kinase, i.e., proto-oncogene tyrosine-protein kinase Src, whose expression and activity are correlated with the malignancy and poor prognosis of numerous types of cancers.77 The oncogenic Y527F mutant of Src can escape from the negative regulation of C-terminal Src kinase (Csk) and exhibits aberrantly high activation levels.78 We thus utilized the Src-Y527F mutant79 and site-specifically incorporated SCouK into the catalytic lysine residue K295 to generate a SCouK-caged Src mutant (Fig. 5C). While cells expressing Src-Y527F-K295SCouK had weak Src autophosphorylation and total tyrosine phosphorylation activities, we observed accumulation of autophosphorylated Src (Src-pY416) and tyrosine-phosphorylated proteins (p-Tyr) in cells irradiated with a 520 nm LED (22 mW cm−2) (Fig. 5D), indicating photoactivation of Src activity leading to downstream signaling. These phosphorylation signals were dependent on irradiation time and reached saturation following irradiation for 60 s (Fig. 5D). As expected, cells expressing Src-Y527F-K295BocK showed no downstream phosphorylation signals under either dark or irradiation conditions (Fig. 5D). Collectively, these results demonstrate that site-specific incorporation of SCouK into key catalytic lysine residues allows for rapid and efficient regulation of kinase activities in live cells.
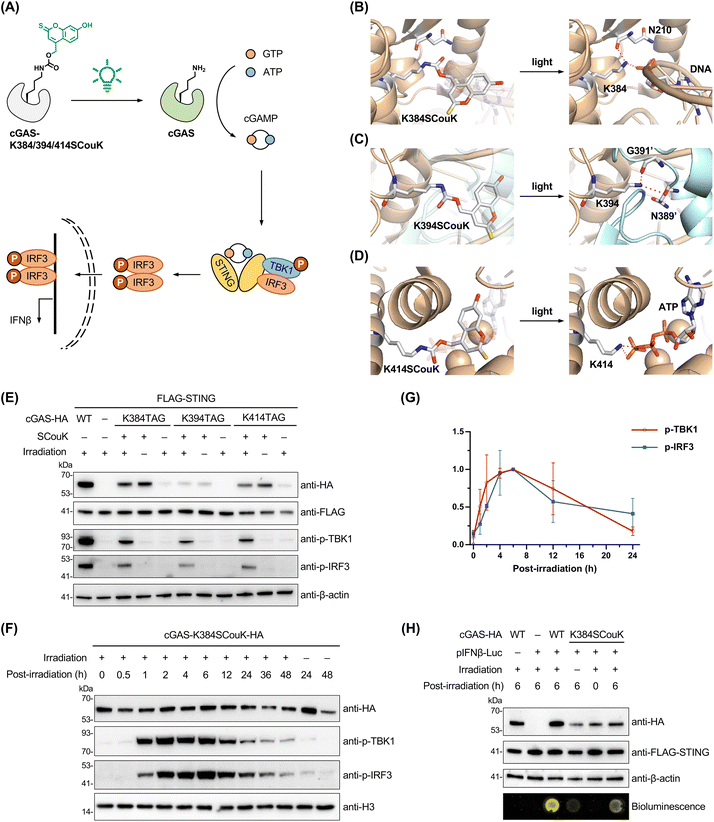
Optical control of cGAS function and the cGAS-STING pathway using SCouK
We next sought to apply SCouK to the optical control of a new signaling enzyme beyond luciferase and kinases. For this, we turned our attention to cyclic GMP-AMP synthase (cGAS) that lies at the core of the cGAS-STING innate immunity signaling pathway.80 As a cytosolic DNA sensor, cGAS assembles on double-stranded DNA (dsDNA), resulting in activation of its catalytic activity and synthesis of cyclic GMP-AMP (cGAMP).81 cGAMP binds to and activates stimulator of interferon genes (STING), which in turn mediates the recruitment of TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3), promoting TBK1 autophosphorylation and IRF3 phosphorylation. The phosphorylated IRF3 translocates to the nucleus and induces the expression of type I interferons (IFNs) and other cytokines (Fig. 6A).82 We envisioned that precise control of cGAS activity could decouple dsDNA-induced cGAS activation from that related to other pathways, such as Mn2+-induced cGAS activation.83 Structural analysis of DNA-bound cGAS suggests that the conserved lysine K384 forms hydrogen bonding with a phosphate group of dsDNA, essential for cGAS-DNA binding (Fig. 6B).84 In addition, previous studies have also shown that the conserved lysine K394 is critical for cGAS dimerization85 (Fig. 6C) and that lysine K414 is involved in binding with ATP (Fig. 6D).86 We thus selected these residues and individually mutated them to the TAG codon in the cGAS gene. The plasmids containing cGAS-K384TAG, cGAS-K394TAG, or cGAS-K414TAG were individually co-introduced with the plasmid expressing SCouKRS and MmPyl-tRNACUA (Fig. S10 and Table S3 in SI) into HEK293T cells stably expressing STING to enable site-specific incorporation of SCouK into cGAS at the designated lysine positions. The co-expression of STING in HEK293T cells was shown to be required for cGAS signaling (Fig. S16 in SI). Western blotting analysis demonstrated that SCouK was efficiently incorporated into the K384, K394, or K414 site, leading to expression of full-length HA-tagged cGAS variants with the same mobility as wild-type (WT) cGAS (Fig. 6E). The faint full-length bands of cGAS-HA observed in the absence of SCouK could be attributed to known low-level near-cognate suppression of the TAG codon.87 TBK1 and IRF3 phosphorylation levels were hardly detected in cells expressing SCouK-modified cGAS in the absence of light illumination (Fig. 6E), indicating that SCouK incorporated at the K384, K394, or K414 site can suppress the activity of cGAS. Notably, much stronger TBK1 and IRF3 phosphorylation signals were observed in cells expressing cGAS-K384SCouK, cGAS-K394SCouK, or cGAS-K414SCouK after green light irradiation (centered at 520 nm, bandwidth ± 19 nm; 22 mW cm−2), suggesting photolysis of SCouK to restore native cGAS and trigger downstream signaling (Fig. 6E).We next focused on cGAS-K384SCouK due to its higher expression level, stronger rescue of cGAS activity, and the critical role of K384 in dsDNA binding. To explore the time dependence of dsDNA-induced cGAS activation and regulation of the cGAS-STING pathway, we performed “pulse-chase” analyses in which cGAS-K384SCouK-expressing cells were briefly irradiated with green light centered at 520 nm (bandwidth ± 19 nm; 22 mW cm−2) and then allowed to recover for different periods in the dark before lysis for western blotting analysis. The results showed that TBK1 phosphorylation burst at about 1 h after irradiation and that IRF3 phosphorylation appeared slightly behind TBK1 phosphorylation (Fig. 6F, G and S17 in SI), which is consistent with the signal transduction axis. Interestingly, both TBK1 and IRF3 phosphorylation signals started to decay at 12 h post-irradiation and almost returned to basal levels after a 2-day recovery period (Fig. 6F and G). These observations suggest the existence of intrinsic regulatory mechanisms to switch off the cGAS-STING signaling pathway and also reveal the time scale of dsDNA-induced cGAS signaling. We further validated the downstream gene expression induced by cGAS activation and IRF3 phosphorylation using the luciferase reporter assay. The luciferase gene in the pIFNβ-Luc plasmid was under the control of an IFNβ promoter and co-transfected with plasmids of cGAS-K384TAG and SCouKRS/MmPyl-tRNACUA in the presence of SCouK. We observed much stronger bioluminescence signals in cells irradiated with green light (520 nm, bandwidth ± 19 nm; 22 mW cm−2) than in non-irradiated cells (Fig. 6H), suggesting that photoactivation of cGAS ultimately leads to IFNβ expression through the cGAS-STING pathway. Together, these results demonstrate that site-specific incorporation of SCouK into the key lysine residue of cGAS allows for precise temporal control and rapid activation of the cGAS-STING signaling pathway in live cells for the first time, providing important insights into the dynamics of the pathway, especially the timing required to initiate and terminate cGAS signaling.
Given the successful demonstrations above, we envision that SCouK could be broadly adapted for the optical control of other proteins. Several considerations should guide the selection of SCouK incorporation sites. In general, SCouK should replace functionally essential lysine residues that are directly involved in catalysis, molecular interactions, or post-translational modifications to transiently silence protein activity, allowing light-triggered restoration of lysine and recovery of protein function. Structural or computational analysis, together with existing biochemical data, can assist in identifying such lysine residues, while empirical screening using functional readouts provides a practical alternative when structural information is limited. Another key consideration is the UAA incorporation efficiency, which depends on the surrounding sequence context88 and can be examined experimentally or predicted computationally.89
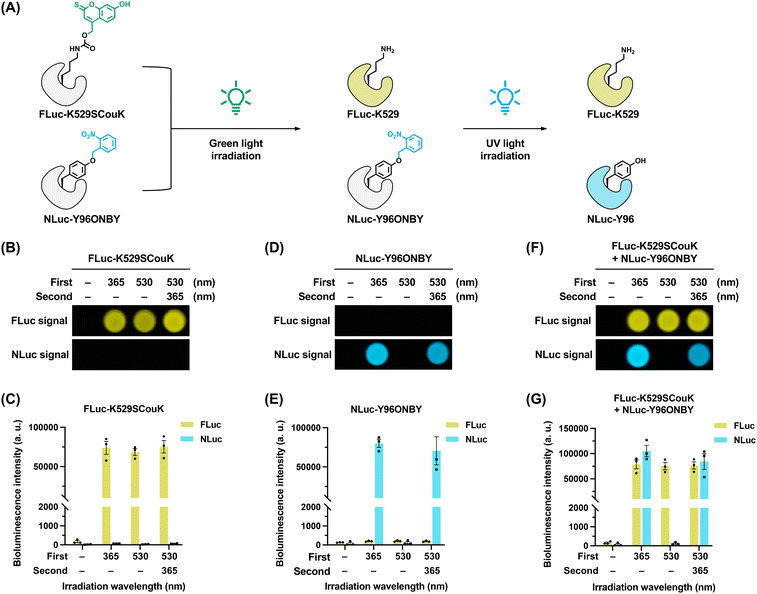
Sequential photoactivation of two proteins using SCouK
We finally explored the capability of SCouK in sequential photoactivation of multiple proteins in a single sample. The broad photoactivation spectrum of SCouK provides great flexibility for various experimental setups, and its long activation wavelengths are orthogonal to UV-responsive photocaged UAAs. However, its wide activation range may also lead to activation at shorter wavelengths, necessitating careful selection of irradiation wavelengths and experimental design in multi-protein studies. In this regard, we chose to cage FLuc with SCouK at the K529 site and NanoLuc luciferase (NLuc)90 with a photolabile tyrosine derivative, o-(2-nitrobenzyl)-tyrosine (ONBY),24,25 at the Y96 site.91 We envisioned that green light irradiation could only decage SCouK to restore FLuc activity and that subsequent UV light irradiation would decage ONBY to switch on NLuc (Fig. 7A). Before the sequential activation experiments, we confirmed the expression of NLuc-Y96ONBY in HEK293T cells by western blotting analysis (Fig. S18A in SI). We again validated that FLuc-K529SCouK can be activated by UV and green light at various wavelengths (e.g., 254 nm, 302 nm, 365 nm, and 520 nm) (Fig. 7B, C and S18B, C in SI). By contrast, NLuc-Y96ONBY was only activated by UV light at 254 nm, 302 nm, or 365 nm, but did not respond to 520 nm green light (Fig. 7D, E and S18B, C in SI). More importantly, we verified that these two luciferase systems are compatible with each other, allowing multiplexing measurements of their activities (Fig. 7B–E). To demonstrate the proof-of-concept for sequential photoactivation of two proteins within a single sample, we employed a simplified system by mixing FLuc-K529SCouK-expressing cells with NLuc-Y96ONBY-expressing cells, rather than performing quadruple transfection in a single cell culture, to ensure sufficient expression of two distinct UAA-modified proteins and to avoid potential cross-interference between the two genetic incorporation systems. These mixed cells were subjected to successive irradiations using green and UV light. Following the first exposure to green light (520 nm, bandwidth ± 19 nm; 22 mW cm−2), we observed robust FLuc signals without any detectable NLuc signals (Fig. 7F and G), indicating that only FLuc-K529SCouK was photoactivated. The subsequent second irradiation with UV light at wavelengths of 365 nm, 254 nm, or 302 nm resulted in strong NLuc signals (Fig. 7F, G and S18B, C in SI), suggesting that NLuc-Y96ONBY was activated following the second light exposure. Therefore, these results demonstrate that SCouK can be sequentially activated with ONBY to enable photocontrol of two distinct proteins in a single live-cell sample. Although these proteins were expressed in separate cell populations, our green-light-responsive SCouK could in future be integrated with other optogenetic tools (such as photo-responsive ligands) or applied to control multiple UAA-modified proteins, or even distinct sites within a single protein, in one cell culture as mutually orthogonal incorporation systems continue to advance, thus offering new opportunities to precisely regulate multiple protein targets and dissect complex cellular mechanisms.Conclusions
In summary, we have developed and genetically encoded a novel photocaged lysine derivative, SCouK, for the optical control of proteins. SCouK is activatable by a wide spectrum of wavelengths, spanning from UV (254 nm, 302 nm, and 365 nm) and near-UV (390 nm) to blue (450 nm) and green (520 nm). In particular, photoactivation of SCouK occurs within 30 s upon exposure to green LED light centered at 520 nm (bandwidth ± 19 nm; 22 mW cm−2), establishing it as the first green-light-responsive UAA with the longest single-photon decaging wavelength reported for genetically encodable photolabile UAAs. What's more, this wavelength is not only more biologically benign but also orthogonal to UV light used to activate current photocaged UAAs. Given the long wavelength and high efficiency for photodecaging, we expect that SCouK will be highly suitable for reducing photodamage and facilitating deep tissue experiments. We have demonstrated the broad utility of SCouK in modulating EGFP fluorescence, luciferase activity, kinase function, and cGAS signaling. Importantly, the site-specific incorporation of SCouK into cGAS enables temporal control and interrogation of the cGAS-STING pathway in live cells for the first time, providing critical insights into its dynamic regulation and signaling. Furthermore, by combing SCouK with a photocaged tyrosine derivative, we have successfully achieved the first sequential photoactivation of two distinct UAAs and their modified proteins within a single live-cell experiment. Collectively, the unique photophysical properties and versatility of SCouK, including green-light-responsiveness, broad and orthogonal decaging wavelengths, and capability for sequential photoactivation of multiple proteins, address the limitations of current UV-responsive lysine-derived UAAs. We believe SCouK significantly enhances our capacity to precisely and simultaneously control and study multiple proteins in complex living systems, opening new opportunities for both functional and therapeutic studies.Author contributions
Li, M.: investigation, writing – original draft; Lu, M.: investigation, writing – original draft; Wang, L.: investigation, methodology; Zhang, Y.: investigation, methodology; Yan, L.: methodology; Wang, S.: methodology; Peng, T.: conceptualization, funding acquisition, project administration, supervision, writing – original draft, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: SI Fig. S1–S18 and Tables S1–S3, experimental procedures including chemical synthesis and biological assays, and characterization of all new compounds. See DOI: https://doi.org/10.1039/d5sc08317f.Acknowledgements
We thank the anonymous reviewers for helpful comments and the Peng lab members for helpful discussion. T.P. acknowledges grant support from Shenzhen Medical Research Fund (B2502022), National Natural Science Foundation of China (22277008), and Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2024SHIBS0004).Notes and references
- S. H. Yun and S. J. J. Kwok, Nat. Biomed. Eng., 2017, 1, 0008 CrossRef CAS PubMed.
- C. Brieke, F. Rohrbach, A. Gottschalk, G. Mayer and A. Heckel, Angew. Chem., Int. Ed., 2012, 51, 8446–8476 CrossRef CAS PubMed.
- N. Ankenbruck, T. Courtney, Y. Naro and A. Deiters, Angew. Chem., Int. Ed., 2018, 57, 2768–2798 CrossRef CAS.
- I. M. Welleman, M. W. H. Hoorens, B. L. Feringa, H. H. Boersma and W. Szymański, Chem. Sci., 2020, 11, 11672–11691 RSC.
- D. Laczi, M. D. Johnstone and C. L. Fleming, Chem.–Asian J., 2022, 17, e202200200 CrossRef CAS.
- Y.-H. Tsai, S. Essig, J. R. James, K. Lang and J. W. Chin, Nat. Chem., 2015, 7, 554–561 CrossRef CAS PubMed.
- J. Broichhagen, A. Damijonaitis, J. Levitz, K. R. Sokol, P. Leippe, D. Konrad, E. Y. Isacoff and D. Trauner, ACS Cent. Sci., 2015, 1, 383–393 CrossRef CAS.
- V. Emiliani, E. Entcheva, R. Hedrich, P. Hegemann, K. R. Konrad, C. Lüscher, M. Mahn, Z.-H. Pan, R. R. Sims, J. Vierock and O. Yizhar, Nat. Rev. Methods Primers, 2022, 2, 55 CrossRef CAS PubMed.
- T. Courtney and A. Deiters, Curr. Opin. Chem. Biol., 2018, 46, 99–107 CrossRef CAS PubMed.
- A. R. Nödling, L. A. Spear, T. L. Williams, L. Y. P. Luk and Y.-H. Tsai, Essays Biochem., 2019, 63, 237–266 CrossRef.
- M. Charette, C. Rosenblum, O. Shade and A. Deiters, Chem. Rev., 2025, 125, 1663–1717 CrossRef CAS.
- J. W. Chin, Nature, 2017, 550, 53 CrossRef CAS PubMed.
- D. D. Young and P. G. Schultz, ACS Chem. Biol., 2018, 13, 854–870 CrossRef CAS.
- M. A. Shandell, Z. Tan and V. W. Cornish, Biochemistry, 2021, 60, 3455–3469 CrossRef CAS PubMed.
- A. Costello, A. A. Peterson, P.-H. Chen, R. Bagirzadeh, D. L. Lanster and A. H. Badran, Chem. Rev., 2024, 124, 11962–12005 CrossRef CAS.
- H. B. Yi, S. Lee, K. Seo, H. Kim, M. Kim and H. S. Lee, Chem. Rev., 2024, 124, 7465–7530 CrossRef CAS PubMed.
- Y. Huang, P. Zhang, H. Wang, Y. Chen, T. Liu and X. Luo, Chem. Rev., 2025, 125, 523–598 CrossRef.
- P. R. Chen, D. Groff, J. Guo, W. Ou, S. Cellitti, B. H. Geierstanger and P. G. Schultz, Angew. Chem., Int. Ed., 2009, 48, 4052–4055 CrossRef CAS PubMed.
- A. Gautier, D. P. Nguyen, H. Lusic, W. An, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2010, 132, 4086–4088 CrossRef CAS PubMed.
- J. Luo, R. Uprety, Y. Naro, C. Chou, D. P. Nguyen, J. W. Chin and A. Deiters, J. Am. Chem. Soc., 2014, 136, 15551–15558 CrossRef CAS PubMed.
- D. P. Nguyen, M. Mahesh, S. J. Elsässer, S. M. Hancock, C. Uttamapinant and J. W. Chin, J. Am. Chem. Soc., 2014, 136, 2240–2243 CrossRef CAS.
- N. Wu, A. Deiters, T. A. Cropp, D. King and P. G. Schultz, J. Am. Chem. Soc., 2004, 126, 14306–14307 CrossRef CAS PubMed.
- W. Ren, A. Ji and H.-w. Ai, J. Am. Chem. Soc., 2015, 137, 2155–2158 CrossRef CAS.
- E. Arbely, J. Torres-Kolbus, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2012, 134, 11912–11915 CrossRef CAS PubMed.
- A. Deiters, D. Groff, Y. Ryu, J. Xie and P. G. Schultz, Angew. Chem., Int. Ed., 2006, 45, 2728–2731 CrossRef CAS.
- J. Luo, J. Torres-Kolbus, J. Liu and A. Deiters, ChemBioChem, 2017, 18, 1442–1447 CrossRef CAS.
- E. A. Lemke, D. Summerer, B. H. Geierstanger, S. M. Brittain and P. G. Schultz, Nat. Chem. Biol., 2007, 3, 769–772 CrossRef CAS.
- X. Ling, Y. Zuo, H. Chen, D. Ji, J. Wang, L. Chang and T. Liu, CCS Chem., 2023, 5, 1301–1307 CrossRef CAS.
- X. Yang, L. Zhao, Y. Wang, Y. Ji, X.-C. Su, J.-A. Ma and W. Xuan, Angew. Chem., Int. Ed., 2023, 62, e202308472 CrossRef CAS.
- X. Zhang, H. Huang, Y. Liu, Z. Wu, F. Wang, X. Fan, P. R. Chen and J. Wang, J. Am. Chem. Soc., 2023, 145, 19218–19224 CrossRef CAS PubMed.
- J. W. Cheung, W. D. Kinney, J. S. Wesalo, M. Reed, E. M. Nicholson, A. Deiters and T. A. Cropp, ChemBioChem, 2023, 24, e202200721 CrossRef CAS.
- A. Gautier, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 2124–2127 CrossRef CAS PubMed.
- J. Liu, J. Hemphill, S. Samanta, M. Tsang and A. Deiters, J. Am. Chem. Soc., 2017, 139, 9100–9103 CrossRef CAS PubMed.
- C. Chou, D. D. Young and A. Deiters, Angew. Chem., Int. Ed., 2009, 48, 5950–5953 CrossRef CAS.
- J. Hemphill, C. Chou, J. W. Chin and A. Deiters, J. Am. Chem. Soc., 2013, 135, 13433–13439 CrossRef CAS.
- S. B. Erickson, R. Mukherjee, R. E. Kelemen, C. J. J. Wrobel, X. Cao and A. Chatterjee, Angew. Chem., Int. Ed., 2017, 56, 4234–4237 CrossRef CAS PubMed.
- T. Bridge, S. A. Shaikh, P. Thomas, J. Botta, P. J. McCormick and A. Sachdeva, Angew. Chem., Int. Ed., 2019, 58, 17986–17993 CrossRef CAS.
- B. Jedlitzke, Z. Yilmaz, W. Dörner and H. D. Mootz, Angew. Chem., Int. Ed., 2020, 59, 1506–1510 CrossRef CAS.
- E. F. Joest, C. Winter, J. S. Wesalo, A. Deiters and R. Tampé, Chem. Sci., 2021, 12, 5787–5795 RSC.
- J. S. Wesalo, Q. Liu, J. Luo, O. Shade and A. Deiters, ChemPhotoChem, 2024, 8, e202300312 CrossRef CAS PubMed.
- S. Wäldchen, J. Lehmann, T. Klein, S. van de Linde and M. Sauer, Sci. Rep., 2015, 5, 15348 CrossRef.
- W. Zhou and A. Deiters, Curr. Opin. Chem. Biol., 2021, 63, 123–131 CrossRef CAS PubMed.
- S. Virdee, P. B. Kapadnis, T. Elliott, K. Lang, J. Madrzak, D. P. Nguyen, L. Riechmann and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 10708–10711 CrossRef CAS.
- W. Ren, A. Ji, M. X. Wang and H.-w. Ai, ChemBioChem, 2015, 16, 2007–2010 CrossRef CAS PubMed.
- S. Wang, X. Wu, Y. Zhang, D. Zhang, B. Xie, Z. Pan, K. Ouyang and T. Peng, Org. Biomol. Chem., 2021, 19, 3469–3478 RSC.
- W. Brown, J. Wesalo, S. Samanta, J. Luo, S. E. Caldwell, M. Tsang and A. Deiters, ACS Chem. Biol., 2023, 18, 1305–1314 CrossRef CAS PubMed.
- J. Icha, M. Weber, J. C. Waters and C. Norden, Bioessays, 2017, 39, 1700003 CrossRef PubMed.
- J. Ma, A. Ripp, D. Wassy, T. Dürr, D. Qiu, M. Häner, T. Haas, C. Popp, D. Bezold, S. Richert, B. Esser and H. J. Jessen, Molecules, 2020, 25, 5325 CrossRef CAS.
- L. Fournier, I. Aujard, T. Le Saux, S. Maurin, S. Beaupierre, J.-B. Baudin and L. Jullien, Chem.–Eur. J., 2013, 19, 17494–17507 CrossRef CAS.
- L. Fournier, C. Gauron, L. Xu, I. Aujard, T. Le Saux, N. Gagey-Eilstein, S. Maurin, S. Dubruille, J.-B. Baudin, D. Bensimon, M. Volovitch, S. Vriz and L. Jullien, ACS Chem. Biol., 2013, 8, 1528–1536 CrossRef CAS.
- D. Manna, B. Maji, S. A. Gangopadhyay, K. J. Cox, Q. Zhou, B. K. Law, R. Mazitschek and A. Choudhary, Angew. Chem., Int. Ed., 2019, 58, 6285–6289 CrossRef CAS PubMed.
- R. Schmidt, D. Geissler, V. Hagen and J. Bendig, J. Phys. Chem. A, 2007, 111, 5768–5774 CrossRef CAS PubMed.
- I. M. Irshadeen, S. L. Walden, M. Wegener, V. X. Truong, H. Frisch, J. P. Blinco and C. Barner-Kowollik, J. Am. Chem. Soc., 2021, 143, 21113–21126 CrossRef CAS PubMed.
- R. T. Michenfelder, F. Pashley-Johnson, V. Guschin, L. Delafresnaye, V. X. Truong, H.-A. Wagenknecht and C. Barner-Kowollik, Adv. Sci., 2024, 11, 2402011 CrossRef CAS.
- J. A. Carroll, F. Pashley-Johnson, M. Klein, T. Stephan, A. K. Pandey, M. Walter, A.-N. Unterreiner and C. Barner-Kowollik, J. Am. Chem. Soc., 2025, 147, 26643–26651 CrossRef CAS.
- W. Wan, J. M. Tharp and W. R. Liu, Biochim. Biophys. Acta, 2014, 1844, 1059–1070 CrossRef CAS.
- T. Peng, T. Das, K. Ding and H. C. Hang, Protein Sci., 2023, 32, e4618 CrossRef CAS.
- N. G. Koch and N. Budisa, Chem. Rev., 2024, 124, 9580–9608 CrossRef CAS PubMed.
- D. L. Dunkelmann and J. W. Chin, Chem. Rev., 2024, 124, 11008–11062 CrossRef CAS PubMed.
- M. Li, F. Wang, L. Yan, M. Lu, Y. Zhang and T. Peng, Chem. Commun., 2022, 58, 10186–10189 RSC.
- M. Li and T. Peng, in Genetically Incorporated Non-Canonical Amino Acids: Methods and Protocols, ed. Y.-H. Tsai and S. J. Elsässer, Springer US, New York, NY, 2023, pp. 55–67, DOI:10.1007/978-1-0716-3251-2_4.
- T. Peng and H. C. Hang, J. Am. Chem. Soc., 2016, 138, 14423–14433 CrossRef CAS PubMed.
- Y. Zhang, Y. Du, M. Li, D. Zhang, Z. Xiang and T. Peng, Angew. Chem., Int. Ed., 2020, 59, 16352–16356 CrossRef CAS PubMed.
- Y. Sun, Y. Chen, Y. Xu, Y. Zhang, M. Lu, M. Li, L. Zhou and T. Peng, Chem. Commun., 2022, 58, 8544–8547 RSC.
- J. S. Spence, R. He, H.-H. Hoffmann, T. Das, E. Thinon, C. M. Rice, T. Peng, K. Chandran and H. C. Hang, Nat. Chem. Biol., 2019, 15, 259–268 CrossRef CAS.
- X. Wu, J. S. Spence, T. Das, X. Yuan, C. Chen, Y. Zhang, Y. Li, Y. Sun, K. Chandran, H. C. Hang and T. Peng, Cell Chem. Biol., 2020, 27, 571–585 CrossRef CAS.
- Y. Li, S. Wang, Y. Chen, M. Li, X. Dong, H. C. Hang and T. Peng, Chem. Commun., 2020, 56, 13880–13883 RSC.
- C. Chen and T. Peng, STAR Protoc., 2020, 1, 100109 CrossRef PubMed.
- Z. J. Zhang, V. A. Pedicord, T. Peng and H. C. Hang, Nat. Chem. Biol., 2020, 16, 95–103 CrossRef CAS.
- E. H. Garst, H. Lee, T. Das, S. Bhattacharya, A. Percher, R. Wiewiora, I. P. Witte, Y. Li, T. Peng, W. Im and H. C. Hang, ACS Chem. Biol., 2021, 16, 844–856 CrossRef CAS PubMed.
- T. Yanagisawa, R. Ishii, R. Fukunaga, T. Kobayashi, K. Sakamoto and S. Yokoyama, Chem. Biol., 2008, 15, 1187–1197 CrossRef CAS.
- D. P. Nguyen, H. Lusic, H. Neumann, P. B. Kapadnis, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2009, 131, 8720–8721 CrossRef CAS.
- N. W. Holton, J. F. Andrews and N. R. Gassman, J. Vis. Exp., 2017, 127, e56265 Search PubMed.
- P. Klán, T. Šolomek, C. G. Bochet, A. Blanc, R. Givens, M. Rubina, V. Popik, A. Kostikov and J. Wirz, Chem. Rev., 2013, 113, 119–191 CrossRef PubMed.
- J. Zhao, S. Lin, Y. Huang, J. Zhao and P. R. Chen, J. Am. Chem. Soc., 2013, 135, 7410–7413 CrossRef CAS PubMed.
- C. J. Caunt, M. J. Sale, P. D. Smith and S. J. Cook, Nat. Rev. Cancer, 2015, 15, 577–592 CrossRef CAS PubMed.
- S. G. Pelaz and A. Tabernero, Oncogene, 2022, 41, 4917–4928 CrossRef CAS.
- T. J. Yeatman, Nat. Rev. Cancer, 2004, 4, 470–480 CrossRef CAS.
- G. Zhang, J. Li, R. Xie, X. Fan, Y. Liu, S. Zheng, Y. Ge and P. R. Chen, ACS Cent. Sci., 2016, 2, 325–331 CrossRef CAS PubMed.
- K.-P. Hopfner and V. Hornung, Nat. Rev. Mol. Cell Biol., 2020, 21, 501–521 CrossRef CAS PubMed.
- Q. Chen, L. Sun and Z. J. Chen, Nat. Immunol., 2016, 17, 1142–1149 CrossRef CAS PubMed.
- M. Motwani, S. Pesiridis and K. A. Fitzgerald, Nat. Rev. Genet., 2019, 20, 657–674 CrossRef CAS.
- Z. Zhao, Z. Ma, B. Wang, Y. Guan, X.-D. Su and Z. Jiang, Cell Rep., 2020, 32(7), 108053 CrossRef CAS.
- F. Civril, T. Deimling, C. C. de Oliveira Mann, A. Ablasser, M. Moldt, G. Witte, V. Hornung and K.-P. Hopfner, Nature, 2013, 498, 332–337 CrossRef CAS.
- X. Zhang, J. Wu, F. Du, H. Xu, L. Sun, Z. Chen, C. A. Brautigam, X. Zhang and Z. J. Chen, Cell Rep., 2014, 6, 421–430 CrossRef CAS PubMed.
- W. Zhou, A. T. Whiteley, C. C. de Oliveira Mann, B. R. Morehouse, R. P. Nowak, E. S. Fischer, N. S. Gray, J. J. Mekalanos and P. J. Kranzusch, Cell, 2018, 174, 300–311 CrossRef CAS PubMed.
- Y. Chemla, E. Ozer, I. Algov and L. Alfonta, Curr. Opin. Chem. Biol., 2018, 46, 146–155 CrossRef CAS PubMed.
- M. Pott, M. J. Schmidt and D. Summerer, ACS Chem. Biol., 2014, 9, 2815–2822 CrossRef CAS PubMed.
- M. D. Bartoschek, E. Ugur, T. A. Nguyen, G. Rodschinka, M. Wierer, K. Lang and S. Bultmann, Nucleic Acids Res., 2021, 49, 62 CrossRef.
- C. G. England, E. B. Ehlerding and W. Cai, Bioconjugate Chem., 2016, 27, 1175–1187 CrossRef CAS PubMed.
- J. Wang, Y. Liu, Y. Liu, S. Zheng, X. Wang, J. Zhao, F. Yang, G. Zhang, C. Wang and P. R. Chen, Nature, 2019, 569, 509–513 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |