 Open Access Article
Open Access ArticlePreparation strategies and biomedical applications of DNA hydrogels
Miaomiao Qiu†
ab,
Jing Wang†
c,
Xinchang Pang
 c,
Dongsheng Liu
c,
Dongsheng Liu
 *de and
Yuanchen Dong
*de and
Yuanchen Dong
 *ab
*ab
aCAS Key Laboratory of Colloid Interface and Chemical Thermodynamics, Beijing National Laboratory for Molecular Sciences, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. E-mail: dongyc@iccas.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cHenan Joint International Research Laboratory of Living Polymerizations and Functional Nanomaterials, Henan Key Laboratory of Advanced Nylon Materials and Application, School of Materials Science and Engineering, Zhengzhou University, Zhengzhou, 450001, China. E-mail: pangxinchang1980@163.com
dThe Hong Kong Polytechnic University, Kowloon, Hong Hum 999077, Hong Kong. E-mail: dongsheng.liu@polyu.edu.hk
ePolyU Shenzhen Research Institute, Hitech Industrial Park, PolyU Base Building, Shenzhen 518057, China
First published on 6th February 2026
Abstract
With the progressive development of DNA nanotechnology and synthetic biology, the applications of DNA have expanded from traditional genetics study to materials science. By employing DNA as a structural framework or cross-linking agent, DNA hydrogels retain a hydrophilic three-dimensional (3D) network structure similar to biological tissues, exhibiting high biocompatibility, programmable responsiveness, and specific recognition functions. In this perspective, we summarize the preparation strategies of DNA hydrogels, analyze their application advantages, and highlight recent advances in areas such as cell culture, drug delivery, and tissue engineering. Finally, we discuss the current challenges in DNA hydrogel development and offer insights into future research directions.
Introduction
Hydrogels are 3D network structures formed by hydrophilic polymers through physical or chemical cross-linking.1 Due to their high water content, excellent biocompatibility and structural similarity to natural biological tissues, hydrogels are widely used in tissue engineering, therapeutic delivery, and biosensing.2 Materials commonly used to construct hydrogels can be classified into two major categories based on their origin: natural materials and synthetic materials. Typical natural materials include gelatin, collagen, hyaluronic acid, and sodium alginate, which exhibit excellent biocompatibility but often suffer from batch-to-batch variability.3 Common synthetic polymers such as polyacrylamide (PAM), polyacrylic acid (PAA), polyvinyl alcohol (PVA), and polyethylene glycol (PEG) offer advantages including high production yield, low cost, and consistent material properties, though they generally demonstrate inferior biocompatibility.4 Consequently, the development of novel hydrogel systems has become a primary focus in hydrogel materials research.Deoxyribonucleic acid (DNA), a natural biomacromolecule, serves as the carrier for genetic information storage and transmission in living organisms. Through the Watson–Crick base-pairing principle, DNA possesses highly selective base recognition capabilities, which confer precise programmability. Through precise sequence design, DNA can be engineered to construct materials with tailored mechanical properties, functionalities, and architectures, offering novel tools and possibilities for biomedical engineering.5,6
In recent years, DNA has demonstrated significant advantages as a biomaterial, leading to extensive exploration in constructing DNA hydrogel systems.7 By employing DNA as a structural scaffold or cross-linker, DNA hydrogels retain the hydrophilic 3D network resembling biological tissues while gaining enhanced biocompatibility, easy functionalization, conditional responsiveness, and specific recognition capabilities. Furthermore, DNA hydrogels offer diverse preparation methods, allowing flexible design according to application requirements. Although challenges remain regarding mechanical strength and production cost, the application potential of DNA hydrogels in therapeutic delivery,8 immunotherapy,9 biosensing,10 and tissue engineering11 is now widely recognized.
In this perspective, we discuss the design strategies, physicochemical properties, and recent advancements in biomedical applications of DNA hydrogels. We further summarize emerging trends and existing challenges in their clinical translation, aiming to provide insights and guidance for future developments in this emerging field.
Design and preparation of DNA hydrogels
Based on their compositional differences, DNA hydrogels can be classified into two categories: hybrid DNA hydrogels and pure DNA hydrogels. In the hybrid DNA hydrogels, DNA serves as the cross-linker and the system usually contains other components as functional materials, e.g., polymer backbone, nano-entities or additional networks. On the other hand, in pure DNA hydrogels, the molecular networks are constructed solely from DNA. The basic types of DNA hydrogels and their corresponding properties are summarized in Table 1.| Type | Molecular network | Cross-linking mechanism | General strength | Biocompatibility | Degradability | Preparation |
|---|---|---|---|---|---|---|
| Hybrid DNA hydrogel | Polymer backbone linked with DNA branches | DNA hybridization | 1–105 Pa; highly adjustable | Depends on the polymer backbone, generally high | Relatively high | Synthesis of the DNA polymer conjugate required |
| DNA hydrogel doped with functional materials | DNA hybridization/physical entanglement | Mostly within the range of 10–103 Pa | Depends on the introduced materials | Relatively high | Easy, requiring only blending/mixing | |
| Double-network DNA hydrogel | DNA hybridization/covalent cross linking | Mostly within the range of 102–103 Pa | Relatively high | Depends on the polymer network | Introduce another monomer into a pre-formed network | |
| Pure DNA hydrogel | Enzymatic ligation | Relies on DNA ligase to form covalent phosphodiester bonds | High, up to 105 Pa | No exogenous toxic substances, relatively high | Relatively high in the pure unmodified system; some DNA modifications, such as phosphorothioate, can reduce the degradability | Need enzymatic reaction |
| Supramolecular DNA hydrogel | Supramolecular self-assembly | Mostly within the range of 102–103 Pa | Easy, mix DNA under specific solutions | |||
| Rolling circle amplification DNA hydrogel | Physical entanglement/DNA hybridization | Low, generally 1–50 Pa | Easy, simply mix the components followed by annealing |
Hybrid DNA hydrogel with DNA cross-linking
In 1996, Nagahara and Matsuda synthesized the first hybrid DNA hydrogel employing DNA as cross-linkers by covalently conjugating acrylate-modified DNA strands to PAM side chains.12 As illustrated in Fig. 1A, they implemented two distinct gelation approaches: by complementary hybridization of two complementary ssDNA linkers grafted onto the polymer backbone and by a free ssDNA strand bridging two non-complementary ssDNA linkers attached to the backbone through simultaneous complementary binding. The DNA-dependent crosslinking mechanism inherently conferred thermo-responsive properties to these hydrogels. Based on the first attempt, diverse stimulus-responsive hydrogels were also engineered through analogous design principles. For example, Willner et al. developed pH-responsive shape-memory hydrogels using a similar architecture (Fig. 1B).13 Two ssDNA strands covalently grafted to a PAM backbone served as cross-linkers, assembling into i-motif structures at pH 5.0 to form stable hydrogels. At pH 8.0, these structures dissociated, triggering hydrogel dissolution into the liquid. Liu et al. constructed an OTA-detecting hybrid DNA hydrogel by functionalizing linear PAM chains with OTA aptamer-encoded DNA strands.14 Upon target recognition, aptamer-OTA binding induced hydrogel network disassembly, releasing encapsulated gold nanoparticles (AuNPs) for visual quantitative detection. The polymer backbones for DNA grafting have also diversified beyond PAM. Biological materials such as polypeptides and proteins have been employed as hydrogel scaffolds. Li et al. utilized (ROP) to graft ssDNA onto polypeptide chains (Fig. 1C), which can form hydrogels through complementary hybridization with dsDNA linkers.15 Furthermore, they substituted the backbone with poly(ethylene glycol)-modified, denatured human serum albumin (HSA).16 Compared to synthetic polypeptides, these natural macromolecules possessed a well-defined primary sequence and secondary structure, offering abundant functionalization sites.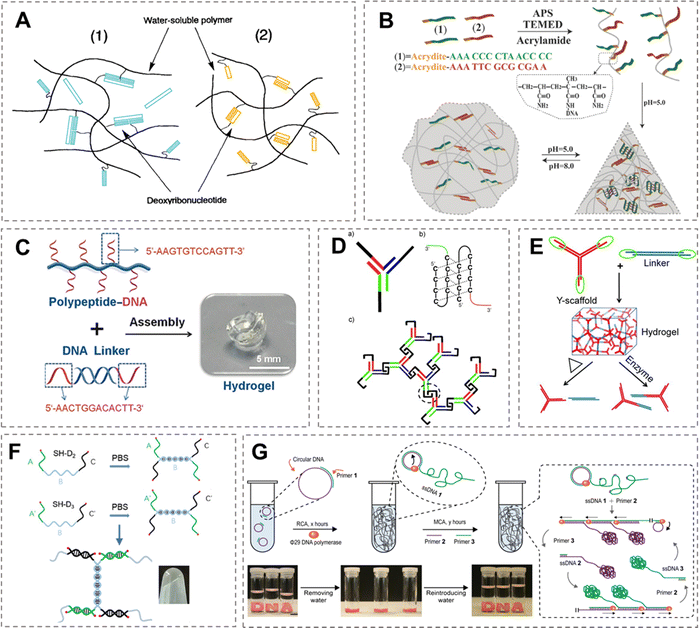 | ||
| Fig. 1 Design and preparation of the DNA hydrogel. (A) Strategies for constructing DNA hybrid hydrogels using oligonucleotides as cross-linkers.12 Permission from Elsevier, copyright 2025. (B) pH-responsive hydrogel synergistically cross-linked by i-motifs.13 Permission from Wiley-VCH, copyright 2025. (C) DNA-peptide hybrid hydrogel formed by grafting ssDNA onto peptide chains and assembling with double-stranded DNA linkers.15 Permission from Wiley-VCH, copyright 2025. (D) Pure DNA supramolecular hydrogel formed via i-motif structures.17 Permission from Wiley-VCH, copyright 2025. (E) Pure DNA supramolecular hydrogel self-assembled from Y-scaffolds and L-linkers.18 Permission from Wiley-VCH, copyright 2025. (F) “H”-shaped pure DNA supramolecular hydrogel cross-linked solely by two types of ssDNA.19 Permission from Wiley-VCH, copyright 2025. (G) Pure DNA hydrogel prepared by multi-primer rolling circle amplification.20 Permission from Springer Nature, copyright 2025. | ||
In addition to grafting DNA onto polymer backbones, hybrid DNA hydrogels can also be formed by incorporating functional materials into the cross-linked network. Through interactions such as electrostatic adsorption, π–π stacking, or coordination bonds, nanomaterials, including AuNPs,21 carbon nanotubes (CNTs),22 graphene oxide (GO),23,24 and lanthanide ions,25 can be attached to DNA strands. As the cross-linked network forms, these functional materials become uniformly dispersed throughout the hydrogel system. Furthermore, to overcome the limitations of single-network structures in mechanical properties and dynamic regulation, a second chemically independent cross-linked network can be constructed within the pure DNA network. These two networks interpenetrate to form a double-network structure. For example, Li et al.26 utilized host–guest interactions between cucurbit[8]uril (CB[8]) and phenylalanine to build an interpenetrating second network in a pure DNA hydrogel system, significantly improving the brittleness and fragility of the single DNA network. Similarly, Cao et al.27 introduced acrylamide/bis-acrylamide monomers into a pure DNA hydrogel, allowing them to diffuse evenly through the DNA network and form a second covalent network in situ, thereby enhancing the mechanical strength of the hydrogel.
Overall, incorporating hybrid components imparts enhanced properties, including superior stability, tunable mechanical strength and controllable luminescence, to DNA hydrogel systems. These advanced functionalities significantly broaden the application prospects of hybrid DNA hydrogels.
Pure DNA hydrogel
Compared to the complex synthesis and modification procedures required for hybrid DNA hydrogels, pure DNA hydrogels demonstrate relative simplicity in both composition and fabrication. Pure DNA hydrogels employ DNA strands as building blocks to form cross-linked networks through either self-assembly or enzymatic ligation. Additionally, physical entanglement and electrostatic interactions can also establish cross-linked networks.In 2006, Luo et al. reported the first pure DNA hydrogel constructed exclusively from single-stranded DNA (ssDNA).28 This system employed an ssDNA self-assembly to generate X-, T-, or Y-shaped DNA building blocks, each featuring variable numbers of complementary sticky ends at their branch terminals. Catalysed by T4 DNA ligase, these structural units interconnected to form stable DNA hydrogels. In 2009, Liu et al. pioneered the construction of pure DNA supramolecular hydrogels by the Y-shaped DNA building blocks (Fig. 1D).17 The system is exclusively composed of DNA strands crosslinked through non-covalent bonding, which employs three partially complementary ssDNA strands assembled under alkaline conditions (pH 8.0) to form Y-shaped units. Upon acidification to pH 5.0, the cytosine-rich termini of adjacent Y-units rapidly dimerized into stable i-motif quadruplex structures. The rigid duplex region at the assembly core prevented intramolecular i-motif folding, directing formation of intermolecular i-motif cross-links that established a 3D hydrogel network. The pH-sensitivity of i-motif structural transitions enabled rapid, reversible sol–gel phase changes in response to pH variation. The same group subsequently refined this design (Fig. 1E) by constructing hydrogels by two modular components: Y-shaped building blocks and L-shaped connectors.18 Jiang et al. designed a ssDNA containing three structural domains, each comprising a self-complementary palindromic sequence. Each individual strand can crosslink with others through these complementary domains, enabling the formation of a solid hydrogel through single-step crosslinking within a single-component system.29 This approach offers notable advantages in compositional simplicity and rapid gelation. In contrast, Du et al. replaced the terminal self-complementary sequences with general sequences and introduced another ssDNA strand containing complementary domains (Fig. 1F).19 Each single strand first forms a dimer via the central self-complementary region, and upon mixing, the dimers undergo secondary crosslinking through the terminal domains. This two-step assembly strategy further enhances the stability and completeness of the crosslinked network. Dirks and Pierce pioneered the hybridization chain reaction (HCR) concept,30 which Wang et al. later extended to the construction of DNA hydrogels.31 Their system employs three DNA strands: introduction of initiator strand I unfolds the hairpin configuration of H1, which then hybridizes with H2 to establish cross-linking. This process proceeds under mild conditions with high assembly fidelity.
DNA hydrogels can alternatively be fabricated through physical entanglement of ultralong DNA strands. As illustrated in Fig. 1G, Luo et al. generated extremely long DNA strands via combined rolling circle amplification (RCA) using Φ29 DNA polymerase and multi-primed chain amplification (MCA).20 These ultralong chains formed hydrogel networks primarily through physically entangled cross-links, supplemented by a limited number of complementary base pairs. As a result, these hydrogels exhibit an extremely low modulus, typically in the range of 1–50 Pa. Remarkably, these hydrogels exhibit unique phase-inversion behaviours, maintaining solid-state integrity in aqueous environments while transitioning to liquid states upon dehydration. On the other hand, DNA hydrogels can alternatively be constructed through entanglement of short DNA assemblies, as demonstrated by Nöll et al., where concentrated short strands self-assembled into linear DNA duplexes whose inter-duplex entanglement established cross-linked networks.32 Furthermore, Liu et al. engineered hydrogels via interpenetrating circular DNA nanostructures, wherein mechanical properties could be tuned by adjusting constituent oligonucleotide rigidity and length.33
In summary, pure DNA hydrogels primarily rely on interbase hydrogen bonding and interchain entanglement for their assembly. Composed exclusively of DNA strands, these systems exhibit exceptional biocompatibility and biodegradability. Consequently, they hold promising applications in biomedical fields such as drug delivery systems, controlled release platforms, and tissue engineering scaffolds.
Physical properties of DNA hydrogels
Capitalizing on the favourable properties of DNA, the mechanical strength and porosity of DNA hydrogels can be precisely tuned within a certain range through rational design of building blocks, adjustment of complementary sequence length, or introduction of additional modifications.34 Furthermore, the reversible hydrogen bonding at the junction sites imparts favourable dynamic properties to DNA hydrogels, such as high permeability, shear-thinning behavior, self-healing capability, and reversible responsiveness to multiple external stimuli.The tunable mechanical strength of DNA hydrogels represents a key advantage for their application in biomedical materials. Liu et al. first proposed the rigid supramolecular DNA hydrogel with tunable mechanical strength, where Y-shaped DNA assemblies and DNA linkers interlock to form a uniform porous structure.18 The backbone distance between crosslinking points was precisely designed to be 84 base pairs, clearly shorter than the persistence length of dsDNA (150 bp).
Building on this work, they further modulated the hydrogel's stiffness by introducing varying numbers of mismatched base pairs within the sticky ends of the two building blocks. They successfully prepared six hydrogel variants with storage moduli ranging from 4.3 kPa to 0.3 kPa (frequency-1 Hz), and utilized this platform to investigate how substrate stiffness influences the differentiation behavior of neural progenitor cells (NPCs).35 Du et al. adopted an alternative strategy by modifying ssDNA terminals with thiol groups.19 Through gradual oxidation, these thiol groups slowly form covalent disulfide bonds, resulting in a hydrogel whose mechanical strength progressively increases over time.
The permeability of DNA hydrogels is a critical property that underpins their utility as biomaterials, as it governs the exchange of nutrients, bioactive factors, therapeutic agents, and metabolic waste. A Y–L DNA supramolecular hydrogel was employed to demonstrate that small molecules can freely diffuse through the DNA hydrogel network, reaching diffusion equilibrium within minutes.36 For larger particles, including macromolecular proteins, Dai et al. proposed a “shutter model” to elucidate the influence of dynamic crosslinking in DNA hydrogels on molecular diffusion.37 According to this model, when the size of a diffusing molecule is smaller than the mesh size of the hydrogel, its diffusion behavior resembles that in free solution. Simultaneously, the dynamic crosslinked network of the DNA hydrogel enables temporary disconnection and rehybridization of junctions, resulting in alternating “open” and “closed” pore states. The “open” state facilitates the diffusion of macromolecules such as proteins. These above studies confirm that DNA hydrogels can effectively support essential material exchange between cells and their surrounding environment.
Benefiting from the reversible nature of DNA base pairing, DNA hydrogels exhibit excellent dynamic properties such as shear-thinning and self-healing behaviors. As the frequency of applied shear increases, the DNA duplexes within the network can undergo reversible dissociation, leading to a rapid decrease in the viscosity of the crosslinked system and demonstrating non-Newtonian fluid characteristics. Upon removal of the shear force, the gel quickly recovers to its original state, endowing DNA hydrogels with injectability and making them suitable as bioinks for 3D bioprinting.15,38 Moreover, when two separated hydrogel pieces are brought into contact, dynamic exchange of complementary base pairing occurs at the interface, allowing them to fuse into an integrated 3D network.39 These dynamic features have enabled extensive research into DNA hydrogels for applications in drug delivery, 3D bioprinting, tissue engineering, and beyond.
Biomedical applications of DNA hydrogels
In biomedical engineering, DNA hydrogels are intelligent materials formed by the self-assembly of programmable DNA sequences. They exhibit high molecular programmability, excellent biocompatibility, and controllable degradability, allowing precise control over cross-linking sites, mechanical properties, degradation kinetics, and functionalization. These features support their applications across scales-from the molecular level to macroscopic systems. Based on their molecular recognition capacity and abundant modification sites, DNA hydrogels have been used for highly sensitive biosensing. Their programmability enables the presentation of antigens or immunomodulatory molecules, making them a promising platform for molecular immunotherapy. Through accurate drug loading and controlled release mechanisms, they provide a targeted drug delivery strategy for nano-medicine. The three-dimensional network also provides a biomimetic microenvironment for cells and allows specific cellular manipulations. Furthermore, by integrating these functions, DNA hydrogels serve as bioactive scaffolds that carry drugs and cells to promote tissue repair and regeneration, thereby advancing tissue engineering.Biosensing
DNA hydrogels demonstrate significant potential in the field of biosensing due to their unique programmable molecular recognition capabilities and stimuli-responsive properties. By converting specific biological stimuli (such as enzymes, nucleic acids, ATP) or non-biological stimuli (such as pH, light, temperature) into physical, chemical, or optical signals, DNA hydrogels enable signal amplification and output through diverse mechanisms.40–42 Their primary functional mechanisms include gel–sol phase transitions,43,44 swelling/deswelling behaviors, optical modulation,45 and changes in mechanical properties.46 This effectively allows the direct transformation of molecular recognition events into detectable signals. This integrated “stimulus-structural transition-signal output” design facilitates highly sensitive and specific detection of trace analytes without requiring additional labels, showcasing considerable value for point-of-care diagnostics and in vivo monitoring.47Sensing strategies based on the gel–sol phase transition represent an early core focus in DNA hydrogel research. The fundamental principle involves target molecules inducing either rupture or reconstruction within the crosslinked network, thereby driving a reversible material transition from the gel state to the sol state. During this process, encapsulated signal molecules can be rapidly released or the properties of the electrode interface altered, resulting in a low-background, high-gain “on–off” signal output. The triggering mechanisms exhibit high programmability, such as (i) utilizing i-motif structures47 or triplex DNA48 to achieve pH-dependent acid–base switching, (ii) incorporating specific enzymatic cleavage sites within crosslinking strands for recognition by proteases or endonucleases,49 (iii) employing strand displacement reactions responsive to specific miRNAs to achieve competitive dissociation of crosslinkers and hydrogel disintegration,50 (iv) exploiting aptamer recognition for targets like ATP or small molecules,51 or leveraging the cleavage of disulfide bonds under reductive environments for specific environmental responses.52 This class of mechanisms has been utilized in various visual and electrochemical detection platforms. For instance, Ma et al. constructed a glucose-responsive sensor using an aptamer-crosslinked DNA hydrogel where target molecule-triggered disintegration released PEG-modified gold nanoparticles (PEG-AuNPs), generating a visible color change for enzyme-free glucose detection with a limit as low as 0.44 mM (Fig. 2A).44 Alternatively, Liu et al. proposed an application where iron-labeled DNA probes crosslinked with DNA strands on a PAM backbone formed a hybrid hydrogel fixed on an electrode surface, where miR-21 presence caused gel disintegration and a subsequent current decrease, enabling sensitive detection at 5 nM (Fig. 2B).53 Furthermore, Zhang et al. integrated the CRISPR/Cas12a system with a rolling circle amplification (RCA)-based DNA hydrogel, where the target nucleic acid activated Cas12a′s trans-cleavage activity, inducing hydrogel disintegration and signal probe release, thereby enabling tri-modal detection (fluorescence, electrochemical, and colorimetric), exemplifying the versatile potential for multimodal detection platform development inherent in this mechanism (Fig. 2C).54
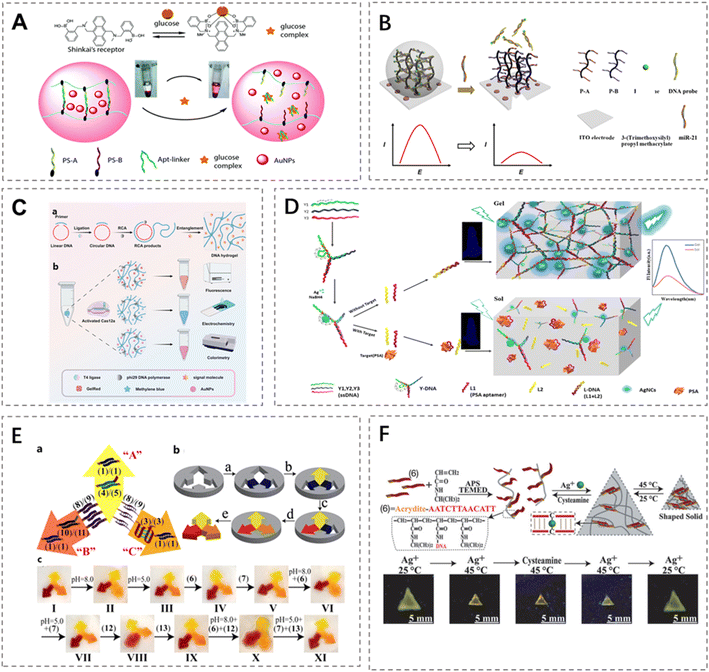 | ||
| Fig. 2 Representative applications of DNA hydrogels in biosensing. (A) A glucose-responsive DNA hydrogel composed of aptamer-crosslinked PAM and embedded AuNPs undergoes structural disassembly upon glucose binding, releasing AuNPs for colorimetric glucose detection.44 Permission from Royal Society of Chemistry, copyright 2025. (B) An electrochemical DNA hydrogel biosensor integrates ferrocene-labeled probes and DNA-grafted polymers for miR-21 detection, where target hybridization triggers probe release and electrochemical signal reduction.53 Permission from Elsevier, copyright 2025. (C) A CRISPR/Cas12a-activated DNA hydrogel prepared via RCA enables multi-modal signal outputs through Cas12a-mediated trans-cleavage of the hydrogel matrix.54 Permission from American Chemical Society, copyright 2025. (D) A PSA-responsive Y-DNA hydrogel with in situ synthesized AgNCs exhibits fluorescence quenching upon target-induced hydrogel dissociation, allowing sensitive PSA quantification.55 Permission from Elsevier, copyright 2025. (E) A programmable shape-memory hydrogel incorporating pH- and strand displacement-responsive DNA motifs enables reversible and region-specific gel-to-quasiliquid transitions for dynamic reconfiguration.56 Permission from American Chemical Society, copyright 2025. (F) A multi-stimuli-responsive DNA-pNIPAM hydrogel utilizes i-motif formation and Ag+ coordination for reversible pH-, ion-, and temperature-triggered sol–gel transitions with excellent mechanical reversibility.57 Permission from WILEY-VCH, copyright 2025. | ||
Distinct from the phase-transition type, swelling/deswelling-based DNA hydrogel sensors achieve macroscopic amplification of molecular recognition events into measurable signals by modulating gel volume changes. In such systems, target molecules drive significant volumetric alterations by influencing crosslinking density, chain conformation, or osmotic pressure; these changes are further transduced into variations such as optical reflection color, Bragg peak shifts, or geometric dimensions. For example, embedding photonic crystals or ordered colloidal crystal arrays within the hydrogel enables direct visual or spectroscopic readout;58 when a target (e.g., a heavy metal ion) binds to recognition groups, altered charge distribution triggers the Donnan effect, causing a shift in lattice spacing and consequently a change in reflection wavelength.59 Alternatively, dynamic hybridization/strand displacement reactions driven by DNA “fuel strands” can induce the extension or cleavage of crosslinkers; reported systems achieve nearly 100-fold volumetric swelling, thereby providing substantial signal amplification.60
In the realm of optical modulation, the excellent optical transparency of DNA hydrogels, combined with functional nanomaterials, endows them with the capability to directly generate and modulate optical signals. Compared to methods relying on macroscopic volume changes, optical signal transduction offers higher sensitivity and faster response times. Primary approaches include fluorescence resonance energy transfer switching and nanomaterial embedding. The donor–acceptor fluorophore pairs are immobilized within the gel network; conformational changes induced by the target molecule alter the distance between them within the 1–10 nm range, enabling fluorescence “on/off” signal switching. Alternatively, incorporating optical nanomaterials such as quantum dots or silver nanoclusters (AgNCs) into the hydrogel allows target response to alter their aggregation state or spatial relationship with quenchers, inducing fluorescence quenching/recovery or aggregation-induced emission (AIE). For instance, in an AgNCs-hydrogel system, the presence of prostate-specific antigen triggers AgNC aggregation, resulting in significant fluorescence enhancement based on the AIE effect, achieving a detection limit as low as 4.4 pg mL−1 and demonstrating exceptional sensitivity for ultra-sensitive detection (Fig. 2D).55
Finally, sensing mechanisms based on changes in mechanical properties offer novel avenues for expanding the functional scope of DNA hydrogels. Through programmable modulation of hydrogel stiffness, elastic modulus, and shape, non-destructive biological monitoring can be achieved. These designs typically rely on orthogonal triggers combined with shape-memory strategies or the integration of thermoresponsive polymers. For example, incorporating both pH-sensitive i-motifs and Ag+-responsive C–Ag+–C pairing units within the same network enables multi-state reversible transitions under distinct stimuli (Fig. 2E).56 Furthermore, copolymerizing thermoresponsive polymers like pNIPAM into the DNA network facilitates a triple phase transition (solution–gel–solid) around 32 °C, where hydrophobic collapse behavior synergistically regulates the hydrogel's mechanical properties alongside DNA conformational changes, thereby enabling dynamic mechanical sensing in complex environments (Fig. 2F).57
Drug delivery
DNA hydrogels are programmable, biodegradable nanomaterials composed solely of DNA strands, whose non-toxic nucleotide degradation products minimize in vivo accumulation.7,61 By tailoring sequence and network architecture, their mechanical properties and cargo-release profiles can be precisely controlled.62 Moreover, intrinsic bioactivity and facile chemical modification enable incorporation of diverse functional groups within a 3D matrix.63 These features establish DNA hydrogels as versatile carrier platforms with significant promise in biomaterials science, particularly for drug delivery applications.64The primary objectives of drug delivery systems are to prolong the drug retention time at target sites, enhance bioavailability, and reduce systemic adverse effects.65 As a new-generation drug carrier, DNA hydrogels enable efficient encapsulation of drugs under mild, biocompatible conditions (often achieving near 100% volumetric pre-loading).66 Beyond chemotherapeutic agents, their compatibility extends to a broad spectrum of bioactive substances, including proteins, nucleic acid therapeutics, and even cells.67,68 Critically, through precise sequence and structural design, DNA hydrogels enable the achievement of accurate spatiotemporal control over drug release kinetics and targeting. For example, Hu et al. provided an acid tolerant, physiological pH responsive DNA hydrogel based on A motif/i motif crosslinking to address the challenges of oral protein delivery (e.g., insulin). The hydrogel protects encapsulated proteins under harsh gastric (pH 1.2) and duodenal (pH 5.0) conditions, dissolving specifically in the small intestine (pH 7.2) for site-specific release (Fig. 3A).69 Fu et al. developed an intelligently responsive DNA nanohydrogel for intracellular mRNA delivery and functional protein expression. It disassembled in the lysosomal acidic environment (pH 4.5–5.0) post-endocytosis, releasing mRNA. Translation efficiency matched commercial LNPs, with superior biocompatibility (Fig. 3B).70
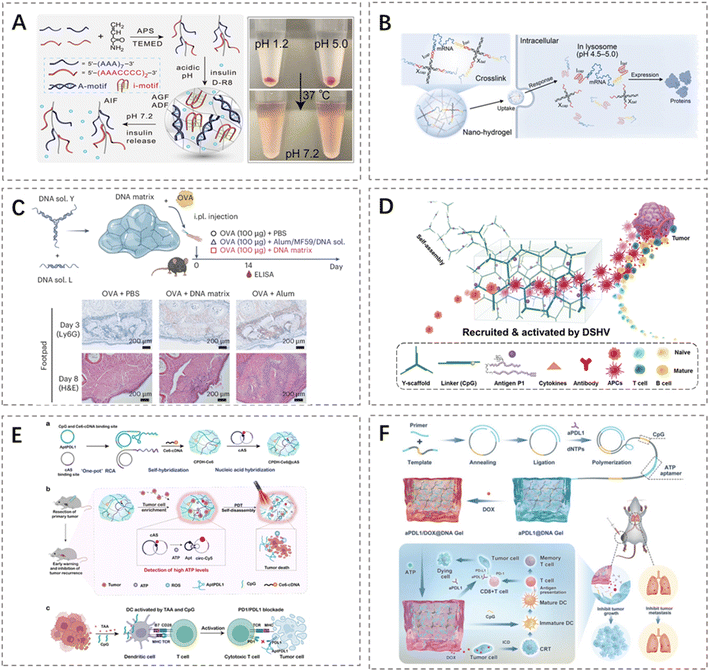 | ||
| Fig. 3 Representative applications of DNA hydrogels in drug delivery and immunotherapy. (A) A pH-responsive DNA hydrogel with A-motif/i-motif dual crosslinking remains stable in acidic gastric environments and disassembles at intestinal pH, enabling targeted oral protein delivery.69 Permission from Wiley-VCH, copyright 2025. (B) An X-shaped DNA scaffold incorporating pH-responsive i-motifs crosslinks mRNA into nanospheres for efficient uptake and pH-triggered cytoplasmic release.70 Permission from Wiley-VCH, copyright 2025. (C) A non-immunostimulatory DNA hydrogel adjuvant enhances both short-term and durable humoral immune responses against SARS-CoV-2 antigens while avoiding autoantibody induction.71 Permission from Springer Nature, copyright 2025. (D) An injectable Y-DNA/CpG supramolecular hydrogel vaccine synergistically activates APCs, inducing potent antitumor immunity with high biocompatibility.72 Permission from Wiley-VCH, copyright 2025. (E) A light-responsive postoperative DNA hydrogel integrating PDL1 aptamer, CpG, and recurrence-sensing modules enables fluorescence-guided relapse detection and laser-triggered immunotherapy.73 Permission from Springer Nature, copyright 2025. (F) A multifunctional DNA hydrogel composed of ATP aptamer/CpG ODN scaffolds co-delivers doxorubicin and anti-PD-L1 antibody, achieving combined chemo-immunotherapy.74 Permission from Wiley-VCH, copyright 2025. | ||
Immunotherapy
The application of DNA hydrogels is particularly transformative in cancer immunotherapy, a field facing challenges like low response rates and immune evasion despite advances in vaccines, checkpoint blockade, and adoptive cell therapy.75–77 DNA hydrogels offer a unique solution as potent carriers of immunostimulatory signals, enabling precise modulation of immune responses. DNA hydrogels can be engineered to locally deliver and sustain immunostimulatory signals (e.g., adjuvants, antigens), mimicking physiological immune microenvironments to potentiate antigen-specific responses.78 This capability is uniquely enabled by the ability to precisely encode bioactive sequences, like cytosine-phosphate-guanine(CpG) motifs, within the DNA scaffold.79 For example, Li et al. constructed an injectable dynamic DNA supramolecular matrix through the self-assembly of five unmodified short-chain ssDNA molecules, serving as a novel vaccine adjuvant platform. This material, as an adjuvant, significantly enhances antigen-specific IgG levels (approximately 1000-fold higher than antigen alone and about 100-fold higher than alum), prolongs the in vivo retention of both the adjuvant and antigen, and undergoes self-disassembly into nanoparticles that promote lymphatic targeting and accumulation. The immunostimulatory effect depends on the TLR9-MyD88 pathway in dendritic cells and is exclusive to right-handed (d-DNA) configurations. In SARS-CoV-2 and Streptococcus pneumoniae infection models, this adjuvant induces high levels of neutralizing antibodies and provides robust protection (Fig. 3C).71 Shao et al. developed an injectable DNA supramolecular hydrogel vaccine (DSHV) that locally enriched CpG motifs to mimic the lymph node microenvironment; this system directly recruits and activates antigen-presenting cells (APCs), thereby eliciting robust immune responses and pronounced antitumor effects (Fig. 3D).72Notably, DNA hydrogels can be engineered as multifunctional platforms that simultaneously incorporate diverse therapeutic modalities, including immunotherapeutic agents, chemotherapeutic drugs, and photosensitizers, within their network architecture.80 Through this precise spatiotemporal control-synergistic combination strategy, DNA hydrogels demonstrate significant potential for advancing personalized and systematic tumor immunotherapy paradigms. For example, Wang et al. engineered an implantable postoperative DNA hydrogel integrating PD-L1 aptamer-mediated capture of recurrent tumor cells, ATP concentration-triggered early warning signaling, and photodynamic immunotherapy for early detection and precise suppression of tumor relapse. This hydrogel concurrently delivers immune checkpoint blockade and adjuvant stimuli to induce systemic antitumor immunity. The system achieved approximately 88.1% inhibition of tumor regrowth and effectively prevented metastatic spread in murine models (Fig. 3E).73 Fan et al. engineered an ultrasoft injectable DNA hydrogel-built on ATP-aptamer scaffolds, polymeric CpG oligonucleotides and co-loaded with anti-PD-L1 and doxorubicin-whose time-programmed release merges chemotoxicity, in situ vaccination, and checkpoint blockade to synergistically inhibit tumor growth, recurrence, and metastasis (Fig. 3F).74
Cell culture and cell capture
The extracellular matrix (ECM) is a dynamic interconnected 3D network residing outside cells, featuring a complex composition. It integrates with cells to maintain tissue integrity while providing mechanical support to embedded cells.81,82 DNA-based networks allow good control over porosity, stiffness, and degradation behavior, thereby providing physicochemical cues reminiscent of native ECM to guide cell adhesion, migration, and differentiation, while also facilitating efficient mass exchange between cells and the matrix.83–86 As a result, DNA hydrogels have emerged as an ideal material platform for mimicking the ECM in biomedical applications.A variety of 3D culture systems have been developed using DNA hydrogels as the substrate to meet different requirements. For example, to enhance DNA hydrogels, Cao et al. constructed a PAM covalent network within a DNA supramolecular hydrogel via in situ polymerization (Fig. 4A).27 This dual-crosslinked network significantly improved tensile and shear strength, offering greater mechanical robustness and enhanced support for encapsulated cells. Li et al. developed a double-network hydrogel using gelatin methacryloyl (GelMA) and DNA, which introduced dynamic functionalities such as stress relaxation, ultimately promoting cell proliferation and osteogenic differentiation.87 Zhu et al. utilized a similar system to investigate how the gel network supports dynamic processes such as tissue mineralization and remodeling during osteogenic differentiation of stem cells.88 Nam et al. functionalized four complementary ssDNA strands with PEG (serving as photo-crosslinking sites) and aptamers (acting as cell-adhesive molecules), respectively, to fabricate multifunctional X-DNA nanostructures (Fig. 4B).89 This system exhibited high mechanical strength, and the aptamer-modified nanostructures further promoted uniform cell distribution and increased cell viability.
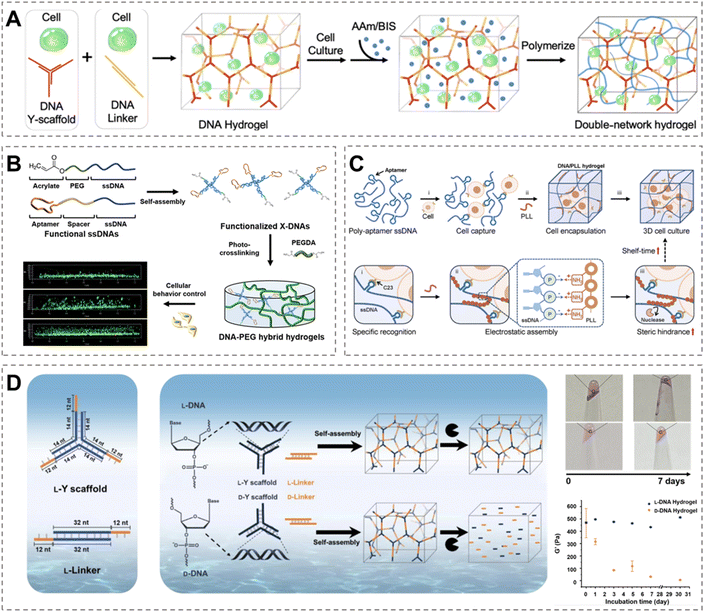 | ||
| Fig. 4 DNA hydrogel system for cell culture. (A) Synthesis of the DNA-PAAm double-network hydrogel. The AAm/BIS monomers diffuse through the DNA supramolecular cross-linked network and construct a second covalent cross-linked network in situ.27 Permission from American Chemical Society, copyright 2025. (B) Synthesis of multifunctional Apt-X-DNA and DNA hybrid hydrogels for 3D cell culture.89 Permission from American Chemical Society, copyright 2025. (C) Cross-linking process of the DNA/PLL hydrogel. The protective coating of PLL increases steric hindrance between DNA and nucleases, extending the service life of the DNA/PLL hydrogel in a 3D cell culture.90 Permission from Wiley-VCH, copyright 2025. (D) The composition of L-DNA and D-DNA hydrogels. The L-DNA hydrogel exhibits significantly higher resistance to nucleases than the D-DNA hydrogel, enabling it to maintain a certain level of mechanical strength over extended periods in the culture medium.91 Permission from Wiley-VCH, copyright 2025. | ||
On the other hand, to increase the bio-stability, Yao et al. proposed a novel stabilization strategy (Fig. 4C) by utilizing poly(L-lysine) (PLL) as a crosslinker.90 The positively charged amino residues present in the PLL coating provide abundant positive charges, which undergo electrostatic adsorption with the negative charges supplied by the phosphate groups of the DNA strands, thereby constructing the cross-linked structure. The steric hindrance introduced by the binding of PLL to DNA impedes the access of nucleases to the DNA strands, thereby enhancing the stability of the DNA/PLL hydrogel and extending its service life to over 15 days. It should be noted that the authors did not investigate the stimuli-responsiveness of the PLL hydrogel, which may be a concern for future applications. Furthermore, though such gels can rapidly transition between gel and sol states when subjected to cyclic strain switching between 1% and 5000%, the effect of the PLL on the dynamic properties in vivo should also be investigated in the future.
In a different approach, Wu et al. employed tetraethylene glycol as a core, conjugating four ssDNA strands to form dendritic DNA molecules, which were subsequently assembled with DNA linkers into a hydrogel network (DDH).92 The DDH network possesses a uniform and densely crosslinked framework, which significantly improves its resistance to endonucleases. Yang et al. reported an L-DNA hydrogel, which exhibits characteristic shear-thinning, self-healing, and thermoresponsive properties (Fig. 4D).91 The left-handed helical conformation of L-DNA confers markedly enhanced biostability to the hydrogel, enabling it to retain its original mechanical strength after immersion in fetal bovine serum (FBS) for over one month. Furthermore, through in vitro cellular assays and a series of inflammatory experiments in mice, the authors confirmed that L-DNA induces almost no inflammatory stimulation, with its induced inflammation level comparable to that of the PBS control group. However, the observation period in mice was limited to six hours. While the six-hour observation period was sufficient to assess acute inflammatory responses, the potential long-term persistence in vivo and its implications for chronic inflammation or immune rejection should be evaluated in the future, which would further strengthen its potential for biomedical applications.
Beyond serving as cell culture substrates, DNA hydrogels show significant promise for the specific capture and release of cells. By readily incorporating targeting modules such as aptamers to construct the hydrogels, these systems can be designed to precisely recognize and isolate specific cell types from complex mixtures. This is primarily achieved through two strategies. One approach involves recruiting target cells after the DNA hydrogel has been formed. For example, Mu et al. developed a pNIPAM-DNA hydrogel featuring a macroporous structure.93 An AS1411 aptamer was incorporated into the DNA sequence to enable specific recognition of cancer cells exhibiting high nucleolin expression, such as MCF-7. Leveraging the thermoresponsive properties of a co-polymerized pNIPAM network, which collapses above its lower critical solution temperature (LCST) and swells below it, the hydrogel enables reversible cell capture and release (Fig. 5A). This system demonstrated high capture efficiency over multiple repeated cycles. In a related approach, Miao et al. constructed a macroporous double-network hydrogel composed of GelMA and DNA using an aqueous bubble-template method.94 The DNA network was functionalized with the Apt19S aptamer, conferring upon the hybrid hydrogel the ability to recruit bone mesenchymal stem cells (BMSCs).
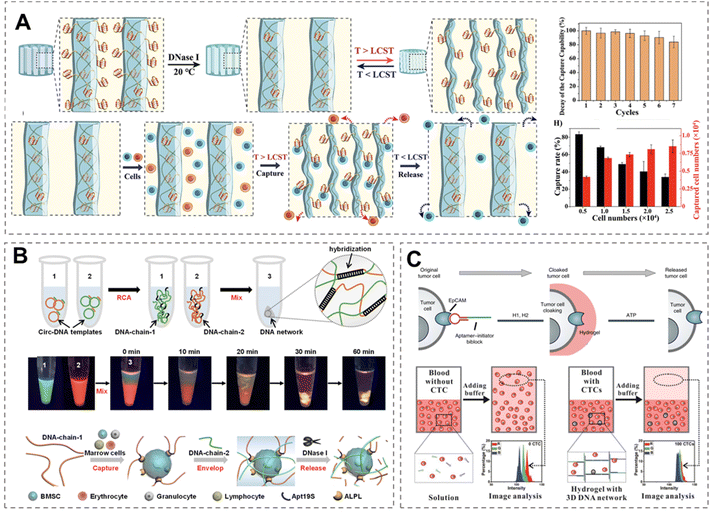 | ||
| Fig. 5 DNA hydrogel system for specific cell capture and release. (A) Schematic illustration of the anisotropic DNA-pNIPAM cryogel for temperature-controlled selective capture and release of target cells.93 Permission from American Chemical Society, copyright 2025. (B) The synthesis process of a physically cross-linked DNA network for stem cell capture.95 Permission from American Chemical Society, copyright 2025. (C) Cell encapsulation based on in situ DNA gelation. The aptamer specifically binds to EpCAM on the cell surface, followed by the triggering of an atcHCR reaction to assemble a DNA hydrogel around the cell. ATP can subsequently bind to the encoded ATP aptamer within the sequence, dissolving the cross-linked structure and releasing the cell.96 Permission from American Chemical Society, copyright 2025. | ||
Alternatively, single-cell encapsulation can be achieved by directly crosslinking ssDNA chains on the cell surface to form a hydrogel network. Yao et al. developed a strategy for capturing cells by directly assembling a DNA hydrogel network on the surface of BMSCs.95 As illustrated in Fig. 5B, two long ssDNA chains were synthesized via RCA. One chain (DNA-chain-1) contained the Apt19S aptamer sequence, which exhibits high affinity toward BMSCs, while the other (DNA-chain-2) was partially complementary to DNA-chain-1. DNA-chain-1 specifically recognized and anchored BMSCs from a mixed cell population. Subsequently, the addition of DNA-chain-2 induced crosslinking with DNA-chain-1 on the cell surface, leading to the formation of an encapsulating DNA hydrogel network around the BMSCs. The captured cells could later be released through enzymatic degradation of the DNA network using nucleases.
Through the same approach, the team also incorporated a PD-1-specific aptamer to enable selective recognition of T cells,97 while functionalizing the network with CpG ODN to activate APCs, thereby achieving a combined immunotherapeutic effect. Li et al. also designed a system in which an EpCAM aptamer was programmed into a DNA strand to act as an initiator for the aptamer-trigger-clamped atcHCR (Fig. 5C).96 When the aptamer specifically recognizes and binds to EpCAM on the target cell surface, it triggers atcHCR, leading to the in situ formation of a hydrogel coating around the cell. This strategy enables precise recognition and isolation of EpCAM overexpressing cells (such as MCF 7) from mixed cell populations, facilitating subsequent cellular analysis.
Tissue engineering
DNA hydrogels represent a novel class of biomaterials capable of forming tunable 3D network structures, providing an ideal platform for tissue engineering applications. Through precise sequence design and controlled molecular assembly, DNA hydrogels offer high controllability over structural mechanical properties, degradation kinetics, and functional site presentation, thereby addressing the diverse regenerative requirements of neural, osseous, cartilaginous, dermal, and vascular tissues.98,99 Notably, their inherent biochemical composition and programmable architecture present distinct advantages in mitigating undesired immune responses. The natural origin and low immunogenicity of DNA could usually reduce the potential for inducing inflammatory or adaptive immune reactions. Although some immunostimulatory sequences such as CpG motifs have been found, these motifs can be easily avoided by the sequence design. The incorporation of antioxidants or anti-inflammatory agents, combined with temporally controlled and stimuli-responsive release profiles, further minimizes prolonged immune exposure. Moreover, DNA hydrogels degrade into biocompatible nucleotides, thereby reducing risks of chronic foreign-body responses. In vivo, DNA hydrogels function not only as delivery vehicles, but also as immunocompatible, injectable or implantable functional scaffolds that actively support biomedical processes including neural,100 bone and cartilage repair,101 wound healing,102 and local angiogenesis.For example, Yuan et al. developed a neural repair scaffold comprising a highly permeable DNA supramolecular hydrogel loaded with neural stem cells for spinal cord regeneration. The construct exploits the intrinsic low immunogenicity and superior permeability of the DNA-based matrix to create a pro-regenerative niche. This design facilitates efficient molecular diffusion, attenuates inflammatory cytokine accumulation, and restrains glial scar formation. By fostering host–graft integration and exerting immunomodulatory effects, the scaffold promotes the re-establishment of functional neural circuits. Consequently, treated animals exhibited significant recovery of hindlimb motor function and detectable electrophysiological responses within eight weeks post-implantation (Fig. 6A).100 In osteochondral regeneration, Yan et al. utilized a tissue engineering strategy in which a pure DNA-based supramolecular hydrogel served as a scaffold for delivering bone marrow-derived BMSCs to treat severe osteoarthritis in a rabbit model. The hydrogel not only shielded MSCs from shear stress during injection but also provided a favorable 3D microenvironment that promoted cell survival, distribution, and chondrogenic differentiation. Moreover, due to its natural DNA composition and biocompatible self-assembly process, the scaffold exhibited low immunogenicity and did not elicit adverse immune responses, as confirmed by reduced inflammatory cytokine levels in the synovial fluid. This integrated scaffold-cell system effectively enhanced cartilage regeneration, demonstrating the potential of DNA hydrogels as immunocompatible, cell-instructive matrices for cartilage tissue engineering (Fig. 6B).103 In the field of skin regeneration, Xiong et al. developed a multilayered regenerative-guidance artificial skin in which programmable DNA hydrogels orchestrate fibroblast behaviors to reconstruct a pro-regenerative microenvironment conducive to scarless tissue remodeling and appendage regeneration. The system integrates an intrinsically biocompatible DNA scaffold with liposome-mediated controlled release to ensure stable biochemical cues, while precise modulation of hydrogel mechanics mitigates aberrant Yes-associated protein mediated mechanotransduction, thereby averting mechanically driven fibrotic remodeling. Importantly, both in vitro and in vivo assessments confirmed that the engineered construct maintains immune homeostasis-promoting reparative macrophage polarization and preventing chronic inflammatory activation-thus enabling the scaffold to facilitate functional neo-tissue formation. Collectively, this regenerative-guidance artificial skin exemplifies a tissue-engineering approach in which biochemical programmability, biomechanical optimization, and immunomodulatory design act synergistically to achieve high-fidelity skin regeneration (Fig. 6C).104 Furthermore, DNA hydrogels can be combined with drug-loaded liposomes or antibacterial/photothermal components, thereby combining bio-responsiveness with anti-infective functions in wound management. For instance, Zhou et al. engineered a photothermally responsive DNA-polyethylenimine hydrogel scaffold integrated with black phosphorus quantum dots for advanced tissue regeneration in diabetic wounds. This multifunctional construct not only exhibits excellent exudate absorption, tissue-adhesive properties, and on-demand antibacterial photothermal capability but also actively orchestrates the regenerative microenvironment. A key feature is its ability to modulate local immune responses by driving macrophage polarization from the pro-inflammatory M1 phenotype toward the pro-regenerative M2 phenotype, thereby redirecting chronic inflammation toward repair and regeneration. Coupled with sustained release of the antioxidant procyanidin B2 to mitigate oxidative stress, the scaffold promotes a cascade of regenerative outcomes, including enhanced angiogenesis, accelerated cutaneous nerve regeneration, and de novo hair follicle formation. To ensure biocompatibility and controlled immune interaction, the design capitalizes on the inherent low immunogenicity of DNA, employs near-infrared-triggered release to minimize prolonged immune exposure, and highlights the importance of optimizing cationic components such as polyethylenimine.105 Notably, Jin et al. employed a shear-thinning and self-healing supramolecular DNA hydrogel for use as an injectable vitreous substitute, which undergoes in situ reassembly into a stable, biomimetic scaffold. Leveraging the low immunogenicity of DNA and its ability to replicate key physicochemical properties of the native vitreous, this system mitigates foreign-body reactions and chronic inflammation, preserves ocular immune privilege, and promotes tissue repair without inducing adverse immune activation (Fig. 6D).106
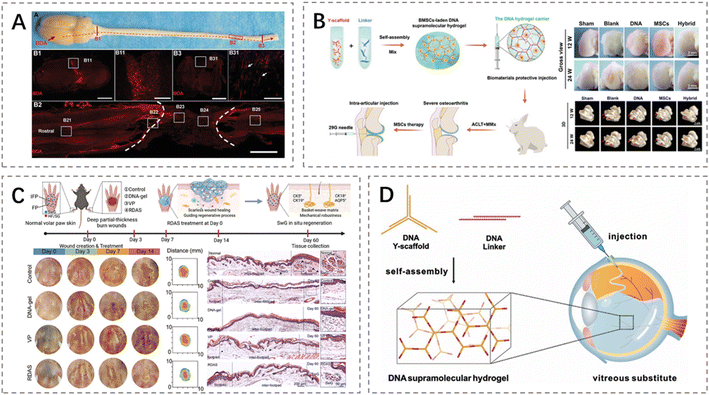 | ||
| Fig. 6 Representative applications of DNA hydrogels in tissue engineering. (A) A highly permeable DNA supramolecular hydrogel loaded with neural stem cells was implanted into spinal cord injury sites, promoting endogenous and transplanted stem cell differentiation to reconstruct neural networks and restore motor function.100 Permission from WILEY-VCH, copyright 2025. (B) An injectable DNA hydrogel carrying bone marrow BMSCs protects them from shear stress and significantly improves osteoarthritis treatment outcomes under mechanically hostile joint conditions.103 Permission from WILEY-VCH, copyright 2025. (C) A bilayer DNA hydrogel with verteporfin-loaded liposomes regulates macrophage polarization and fibroblast regeneration, enabling scar-free wound healing and restoration of functional skin appendages.104 Permission from WILEY-VCH, copyright 2025. (D) A self-assembled DNA supramolecular hydrogel mimicking vitreous properties exhibits high mechanical strength, shear-thinning, and long-term biocompatibility, effectively maintaining retinal structure and intraocular pressure in vivo.106 Permission from Wiley-VCH, copyright 2025. | ||
To summarize, the multifunctionality of DNA hydrogels stems from their highly programmable molecular scaffold and readily functionalizable chemical sites. Through precise sequence design and the integration of modular functional elements, these materials are endowed with the ability to dynamically respond to biological signals, execute complex instructions, and ultimately achieve predefined biological goals. However, their clinical translation faces several common challenges, including long-term stability in complex physiological environments, scalable synthesis and cost control, tissue-specific delivery in vivo, precise assessment and modulation of material-host immune interactions, and the establishment of standardized evaluation systems bridging in vitro characterization and in vivo functional validation. Future research will increasingly focus on developing logic-gated systems capable of multimodal responsiveness and autonomous feedback. By fostering interdisciplinary integration, spanning artificial intelligence, synthetic biology, living cell therapeutics, and advanced manufacturing technologies, efforts will be directed toward establishing a closed-loop research framework from sequence–structure–function theoretical design to in vivo functional validation. The ultimate goal is to evolve DNA hydrogels from “smart materials” into bioinspired interfaces capable of deep information exchange with biological systems, thereby providing actionable technological pathways and methodological foundations for precision medicine and regenerative medicine.
Conclusions and outlook
Since the first construction of 3D gel networks with DNA molecules, DNA hydrogels have undergone multiple developmental stages, from conceptual establishment and performance optimization to functional integration. Owing to their physicochemical properties resembling the native extracellular matrix, along with unique advantages such as programmability, precise molecular recognition, stimuli-responsiveness, self-healing capabilities, and ease of functionalization, DNA hydrogels have emerged as an ideal platform for engineering smart biomaterials. They demonstrate significant potential in a wide range of applications including tissue engineering, drug delivery, cell culture, and biosensing. In recent years, researchers have further enhanced the mechanical properties and functionality of DNA hydrogels through strategies such as chemical modification of DNA strands, rational design of crosslinking modules, and hybridization with various functional materials, greatly expanding their utility in biomedical applications.Despite the significant application potential of DNA hydrogels, their development thus far remains confined to laboratory settings, with no approved clinical trials to date. This limitation stems from multiple challenges, including a narrow range of mechanical strength, extremely low production efficiency and high costs of purification and modification, which are currently inadequate to meet the demands of clinical trials, and the long-term biological safety of these materials has yet to be fully validated.
Currently, most commercially available synthetic DNA is produced and purified in laboratories, with quantities typically measured in micromoles or millimoles. In contrast, synthetic prepolymers can be manufactured on a scale of kilograms or even tons. This situation indicates that DNA production lags far behind that of synthetic polymers. Moreover, many DNA modification strategies further increase the complexity and cost of hydrogel preparation. For example, L-DNA contains deoxyribose, which is a mirror image of D-DNA, so conventional commercial DNA synthesis monomers cannot be used. Although many teams optimized the synthesis route for these mirror-image monomers and achieved a scale of tens of grams, the high monomer consumption in DNA synthesis still makes L-DNA very expensive. As a result, it remains difficult to apply in studies involving medium or large animal models. Overall, the synthesis of DNA, including modified or functionalized DNA, faces major limitations. These include high costs from expensive raw materials, specialized equipment, and controlled synthesis environments, as well as multi-step processes and low yields. Together, these factors significantly hinder large-scale production.
Furthermore, the mechanical properties of DNA hydrogels remain inadequate, which limited their applications in tissue engineering. Different biological tissues exhibit elastic moduli that span several orders of magnitude. Compared to synthetic polymers which possess stable mechanical properties and a modulus span of up to several million Pascals, the modulus of DNA hydrogels is typically limited to the range of hundreds to thousands of Pascals. Although this stiffness confers advantages for repairing soft tissues in vivo, such as skins, muscles, and nerves, it falls significantly short for high-strength applications. Modifications or adulterations could substantially enhance their strength; however, such approaches often raise additional safety concerns and increase costs. This considerable mechanical property gap has also severely hampered the expansion of their application scope. Thirdly, DNA hydrogels face challenges in biological stability and long-term safety. Native DNA is susceptible to enzymatic degradation by nucleases, and this vulnerability also extends to DNA hydrogels. In certain applications, such as controlled drug release or the construction of biodegradable tissue engineering scaffolds, this degradability can be advantageous. However, in contexts requiring long term structural integrity, such as in prolonged cell culture systems, it becomes a significant drawback. More importantly, studies have shown that fragments resulting from DNA degradation may potentially trigger immune responses, further raising concerns about medical applicability. Fourth, the response kinetics of current stimulus responsive DNA hydrogels, including those activated by light, enzymes, or magnetic fields, remain relatively slow. The delayed structural reorganization hinders their utility in real time regulation and precision medicine applications. We have provided a comparative table summarizing the respective advantages and disadvantages of natural hydrogels, synthetic polymer hydrogels, and DNA hydrogels, which more visually highlights the strengths of DNA hydrogels as well as their limitations in clinical applications (Table 2).
| Type | Common materials | Main advantages | Main disadvantages | Preparation difficulty | Commercialized products |
|---|---|---|---|---|---|
| Natural hydrogels | Collagen, hyaluronic acid, gelatin, fibrin, alginate, chitosan, etc. | Good biocompatibility | Weak mechanical strength | Easy raw material acquisition, difficult purification or modification | Hyaluronic acid gels: dermal fillers, e.g., Juvéderm, Restylane |
| Cell recognition sites | Poor stability; potential pathogen immunogenicity | ||||
| Clear metabolic pathways | High batch-to-batch variability | ||||
| Inherent biological functions | |||||
| Synthetic hydrogels | PEG, PAA, PAM, PVA, etc. | Stable mechanical properties | Low bioactivity | Relatively simple with high commercial availability | PEG: contact lenses; PPA: core moisture-absorbing material in wound dressings, e.g., Aquacel |
| High batch consistency | Poor dynamic response | ||||
| Easy to modify | |||||
| DNA hydrogels | DNA strands | Good programmability; dynamic responsiveness | High cost | Easy manipulation | N/A |
| High biocompatibility | Low yield | ||||
| Molecular recognition capability | Short service life; easily degradable |
To address these challenges, future development of DNA hydrogels requires the assistance of many relevant directions. First, ongoing optimization of DNA synthesis and modification strategies is needed to reduce production costs. The currently mature phosphoramidite solid-phase synthesis system yields relatively small quantities per run, sufficient for experimental studies but inadequate for large-scale animal experiments or clinical research with higher material demands. Therefore, breakthroughs can be sought in instrument design or synthetic route optimization, such as developing high-throughput or large-scale production equipment, and refining DNA synthesis processes to lower costs and improve efficiency. Second, simpler and more efficient hydrogel assembly techniques should be developed, including enzyme-free self-assembly and improved RCA processes. Third, integrating artificial intelligence (AI) into the design of DNA hydrogels can aid in sequence design and performance prediction. Currently, many AI models are already used to predict non-DNA hydrogel properties107 such as assembly structure and modulus,108 which benefit the prediction of drug loading and release, the design of hydrogel shape and functions for wound healing, and other medical assistance schemes. However, such AI models have not been currently utilized in DNA hydrogels. With the aid of such artificial intelligence, it can be anticipated that large-scale data analysis, predictive study, and algorithm optimization can be conducted efficiently in the future. For example, in drug delivery systems, it would be possible to design and program optimal DNA chain lengths or cross-linking structures based on the properties of drugs. It will also provide a facile tool to optimize the pore size of the cross-linked network to achieve optimal drug loading efficiency. Alternatively, integrated applications such as predicting drug release profiles based on DNA degradation rates and developing smart sensors leveraging the molecular recognition capabilities of DNA may significantly enhance the development efficiency of DNA hydrogels. Moreover, multifunctional integrated DNA hydrogel systems would be developed rapidly. For instance, incorporating bioactive factors, functional nanomaterials, and stimuli-responsive components into the cross-linked network enables synergistic control over mechanical, electrical, and biological guidance properties, along with degradation behavior. Finally, greater emphasis must be placed on clinical translation. Interdisciplinary collaborations should be strengthened to conduct systematic in vitro and in vivo evaluations of biosafety and efficacy, advancing DNA hydrogels as viable biomaterials in practical applications. These innovations could offer novel solutions for real-world applications in precision medicine and regenerative therapies.
In summary, as an emerging biomaterial, DNA hydrogels-though facing multiple challenges-exhibit considerable potential for development. With continuous progress and increasing interdisciplinary integration across fields such as materials science, chemistry, biology, and medicine, it is expected that through multidisciplinary collaboration and technological convergence, current limitations can be overcome, enabling a successful transition from laboratory research to clinical practice.
Author contributions
All authors contributed to the writing and revision of the manuscript and have approved the final version of the perspective.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included, and no new data were generated or analysed as part of this perspective.Acknowledgements
The authors would like to acknowledge the National Basic Research Plan of China (2024YFA1308500, 2023YFA0915201), Beijing Natural Science Foundation (JQ24007, L254021), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0960000), National Natural Science Foundation of China (22477122), and the Open Project of State Key Laboratory of Supramolecular Structure and Materials (sklssm202506).References
- O. Wichterle and D. LÍM, Hydrophilic Gels for Biological Use, Nature, 1960, 185, 117–118 CrossRef.
- D. Seliktar, Designing Cell-Compatible Hydrogels for Biomedical Applications, Science, 2012, 336, 1124–1128 CrossRef CAS PubMed.
- L. Zhao, Y. Zhou, J. Zhang, H. Liang, X. Chen and H. Tan, Natural Polymer-Based Hydrogels: From Polymer to Biomedical Applications, Pharmaceutics, 2023, 15, 2514 CrossRef CAS PubMed.
- J. Thiele, Y. Ma, S. M. C. Bruekers, S. Ma and W. T. S. Huck, 25th Anniversary Article: Designer Hydrogels for Cell Cultures: A Materials Selection Guide, Adv. Mater., 2014, 26, 125–148 CrossRef CAS PubMed.
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- D. Yang, M. R. Hartman, T. L. Derrien, S. Hamada, D. An, K. G. Yancey, R. Cheng, M. Ma and D. Luo, DNA Materials: Bridging Nanotechnology and Biotechnology, Acc. Chem. Res., 2014, 47, 1902–1911 CrossRef CAS PubMed.
- J. Li, L. Mo, C.-H. Lu, T. Fu, H.-H. Yang and W. Tan, Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications, Chem. Soc. Rev., 2016, 45, 1410–1431 RSC.
- F. Mo, K. Jiang, D. Zhao, Y. Wang, J. Song and W. Tan, DNA hydrogel-based gene editing and drug delivery systems, Adv. Drug Deliv. Rev., 2021, 168, 79–98 Search PubMed.
- J. Gačanin, C. V. Synatschke and T. Weil, Biomedical Applications of DNA-Based Hydrogels, Adv. Funct. Mater., 2020, 30, 1906253 CrossRef.
- L. Xi, Y. Shang, Z. Wang, J. Wang, Q. Wu, Y. Shen and Y. Ding, Programmable DNA hydrogels for biosensing and point-of-care test, Coord. Chem. Rev., 2024, 518, 35 Search PubMed.
- C. Chen, J. Zhou, J. Chen and H. Liu, Advances in DNA Supramolecular Hydrogels for Tissue Engineering, Macromol. Biosci., 2022, 22, 2200152 CrossRef CAS PubMed.
- S. Nagahara and T. Matsuda, Hydrogel formation via hybridization of oligonucleotides derivatized in water-soluble vinyl polymers, Polym. Gels Netw., 1996, 4, 111–127 CrossRef CAS.
- W. Guo, C.-H. Lu, R. Orbach, F. Wang, X.-J. Qi, A. Cecconello, D. Seliktar and I. Willner, pH-Stimulated DNA Hydrogels Exhibiting Shape-Memory Properties, Adv. Mater., 2015, 27, 73–78 CrossRef CAS PubMed.
- R. Liu, Y. Huang, Y. Ma, S. Jia, M. Gao, J. Li, H. Zhang, D. Xu, M. Wu, Y. Chen, Z. Zhu and C. Yang, Design and Synthesis of Target-Responsive Aptamer-Cross-linked Hydrogel for Visual Quantitative Detection of Ochratoxin A, ACS Appl. Mater. Interfaces, 2015, 7, 6982–6990 CrossRef CAS PubMed.
- C. Li, A. Faulkner-Jones, A. R. Dun, J. Jin, P. Chen, Y. Xing, Z. Yang, Z. Li, W. Shu, D. Liu and R. R. Duncan, Rapid Formation of a Supramolecular Polypeptide–DNA Hydrogel for In Situ Three-Dimensional Multilayer Bioprinting, Angew. Chem., Int. Ed., 2015, 54, 3957–3961 CrossRef CAS PubMed.
- Y. Wu, C. Li, F. Boldt, Y. Wang, S. L. Kuan, T. T. Tran, V. Mikhalevich, C. Förtsch, H. Barth, Z. Yang, D. Liu and T. Weil, Programmable protein–DNA hybrid hydrogels for the immobilization and release of functional proteins, Chem. Commun., 2014, 50, 14620–14622 RSC.
- E. Cheng, Y. Xing, P. Chen, Y. Yang, Y. Sun, D. Zhou, L. Xu, Q. Fan and D. Liu, A pH-Triggered, Fast-Responding DNA Hydrogel, Angew. Chem., Int. Ed., 2009, 48, 7660–7663 CrossRef CAS PubMed.
- Y. Xing, E. Cheng, Y. Yang, P. Chen, T. Zhang, Y. Sun, Z. Yang and D. Liu, Self-Assembled DNA Hydrogels with Designable Thermal and Enzymatic Responsiveness, Adv. Mater., 2011, 23, 1117–1121 CrossRef CAS PubMed.
- X. Du, Y. Xing, Y. Li, M. Cao, J. Wu, G. Dong, Z. Shi, X. Wei, M. Qiu, J. Gao, Y. Xu, H. Xu, D. Liu and Y. Dong, Gradually Self-Strengthen DNA Supramolecular Hydrogels, Macromol. Rapid Commun., 2024, 45, 2400177 CrossRef CAS PubMed.
- J. B. Lee, S. Peng, D. Yang, Y. H. Roh, H. Funabashi, N. Park, E. J. Rice, L. Chen, R. Long, M. Wu and D. Luo, A mechanical metamaterial made from a DNA hydrogel, Nat. Nanotechnol., 2012, 7, 816–820 CrossRef CAS PubMed.
- B. Liu, J. Sun, J. Zhu, B. Li, C. Ma, X. Gu, K. Liu, H. Zhang, F. Wang, J. Su and Y. Yang, Injectable and NIR-Responsive DNA–Inorganic Hybrid Hydrogels with Outstanding Photothermal Therapy, Adv. Mater., 2020, 32, 2004460 CrossRef CAS PubMed.
- E. Cheng, Y. Li, Z. Yang, Z. Deng and D. Liu, DNA-SWNT hybrid hydrogel, Chem. Commun., 2011, 47, 5545–5547 RSC.
- L. Sun, N. Hu, J. Peng, L. Chen and J. Weng, Ultrasensitive Detection of Mitochondrial DNA Mutation by Graphene Oxide/DNA Hydrogel Electrode, Adv. Funct. Mater., 2014, 24, 6905–6913 CrossRef CAS.
- Y. Xu, Q. Wu, Y. Sun, H. Bai and G. Shi, Three-Dimensional Self-Assembly of Graphene Oxide and DNA into Multifunctional Hydrogels, ACS Nano, 2010, 4, 7358–7362 CrossRef CAS PubMed.
- S. Yang, X. Pan, J. Tang, C. Yao and D. Yang, Lanthanide-DNA supramolecular hydrogels with tunable and responsive luminescence, Sci. China Technol. Sci., 2022, 65, 1043–1051 CrossRef CAS.
- C. Li, M. J. Rowland, Y. Shao, T. Cao, C. Chen, H. Jia, X. Zhou, Z. Yang, O. A. Scherman and D. Liu, Responsive Double Network Hydrogels of Interpenetrating DNA and CB[8] Host–Guest Supramolecular Systems, Adv. Mater., 2015, 27, 3298–3304 Search PubMed.
- T. Cao, H. Jia, Y. Dong, S. Gui and D. Liu, In Situ Formation of Covalent Second Network in a DNA Supramolecular Hydrogel and Its Application for 3D Cell Imaging, ACS Appl. Mater. Interfaces, 2020, 12, 4185–4192 CrossRef CAS PubMed.
- S. H. Um, J. B. Lee, N. Park, S. Y. Kwon, C. C. Umbach and D. Luo, Enzyme-catalysed assembly of DNA hydrogel, Nat. Mater., 2006, 5, 797–801 CrossRef CAS PubMed.
- H. Jiang, V. Pan, S. Vivek, E. R. Weeks and Y. Ke, Programmable DNA Hydrogels Assembled from Multidomain DNA Strands, ChemBioChem, 2016, 17, 1156–1162 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Triggered amplification by hybridization chain reaction, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- J. Wang, J. Chao, H. Liu, S. Su, L. Wang, W. Huang, I. Willner and C. Fan, Clamped Hybridization Chain Reactions for the Self-Assembly of Patterned DNA Hydrogels, Angew. Chem., Int. Ed., 2017, 56, 2171–2175 Search PubMed.
- T. Nöll, H. Schönherr, D. Wesner, M. Schopferer, T. Paululat and G. Nöll, Construction of Three-Dimensional DNA Hydrogels from Linear Building Blocks, Angew. Chem., Int. Ed., 2014, 53, 8328–8332 Search PubMed.
- H.-Y. Jia, J.-Z. Shi, Y. Shao and D.-S. Liu, DNA hydrogels formed of bended DNA scaffolds and properties study, Chin. J. Polym. Sci., 2017, 35, 1307–1314 CrossRef CAS.
- Y. Li, R. Chen, B. Zhou, Y. Dong and D. Liu, Rational Design of DNA Hydrogels Based on Molecular Dynamics of Polymers, Adv. Mater., 2024, 36, 2307129 CrossRef CAS PubMed.
- B. Zhou, B. Yang, Q. Liu, L. Jin, Y. Shao, T. Yuan, Y.-n. Zhang, C. Wang, Z. Shi, X. Li, Y. Pan, N. Qiao, J.-F. Xu, Y. R. Yang, Y. Dong, L. Xu, S. Gui and D. Liu, Effects of Univariate Stiffness and Degradation of DNA Hydrogels on the Transcriptomics of Neural Progenitor Cells, J. Am. Chem. Soc., 2023, 145, 8954–8964 CrossRef CAS PubMed.
- J. Jin, Y. Xing, Y. Xi, X. Liu, T. Zhou, X. Ma, Z. Yang, S. Wang and D. Liu, A Triggered DNA Hydrogel Cover to Envelop and Release Single Cells, Adv. Mater., 2013, 25, 4714–4717 CrossRef CAS PubMed.
- X. Dai, Z. Zhu, Y. Li, B. Yang, J.-F. Xu, Y. Dong, X. Zhou, L.-T. Yan and D. Liu, “Shutter” Effects Enhance Protein Diffusion in Dynamic and Rigid Molecular Networks, J. Am. Chem. Soc., 2022, 144, 19017–19025 CrossRef CAS PubMed.
- J. Shi, Y. Wan, H. Jia, G. Skeldon, D. Jan Cornelissen, K. Wesencraft, J. Wu, G. McConnell, Q. Chen, D. Liu and W. Shu, Printing Cell Embedded Sacrificial Strategy for Microvasculature using Degradable DNA Biolubricant, Angew. Chem., Int. Ed., 2025, 64, e202417510 CrossRef CAS PubMed.
- Y. Wang, Y. Shao, X. Ma, B. Zhou, A. Faulkner-Jones, W. Shu and D. Liu, Constructing Tissuelike Complex Structures Using Cell-Laden DNA Hydrogel Bricks, ACS Appl. Mater. Interfaces, 2017, 9, 12311–12315 CrossRef CAS PubMed.
- F. Qi, H. Li, Y. Wang and C. Ding, Responsive DNA hydrogels: design strategies and prospects for biosensing, Chem. Commun., 2024, 60, 10231–10244 RSC.
- F. Li, D. Lyu, S. Liu and W. Guo, DNA hydrogels and microgels for biosensing and biomedical applications, Adv. Mater., 2020, 32, 1806538 CrossRef CAS PubMed.
- S. Khajouei, H. Ravan and A. Ebrahimi, DNA hydrogel-empowered biosensing, Adv. Colloid Interface Sci., 2020, 275, 102060 CrossRef CAS PubMed.
- Z. Zhu, Z. Guan, S. Jia, Z. Lei, S. Lin, H. Zhang, Y. Ma, Z. Q. Tian and C. J. Yang, Au@ Pt nanoparticle encapsulated target-responsive hydrogel with volumetric bar-chart chip readout for quantitative point-of-care testing, Angew. Chem., Int. Ed., 2014, 53, 12503–12507 Search PubMed.
- Y. Ma, Y. Mao, Y. An, T. Tian, H. Zhang, J. Yan, Z. Zhu and C. J. Yang, Target-responsive DNA hydrogel for non-enzymatic and visual detection of glucose, Analyst, 2018, 143, 1679–1684 RSC.
- W. Guo, R. Orbach, I. Mironi-Harpaz, D. Seliktar and I. Willner, Fluorescent DNA hydrogels composed of nucleic acid-stabilized silver nanoclusters, Small, 2013, 9, 3748–3752 Search PubMed.
- S. R. Deshpande, R. Hammink, R. K. Das, F. H. Nelissen, K. G. Blank, A. E. Rowan and H. A. Heus, DNA-Responsive Polyisocyanopeptide Hydrogels with Stress-Stiffening Capacity, Adv. Funct. Mater., 2016, 26, 9075–9082 CrossRef CAS.
- S. Iqbal, F. Ahmed and H. Xiong, Responsive-DNA hydrogel based intelligent materials: Preparation and applications, Chem. Eng. J., 2021, 420, 130384 Search PubMed.
- Y. Hu, Y. Ke and I. Willner, A pH-Cascaded DNA Hydrogel Mediated by Reconfigurable A-motif Duplex, i-Motif Quadruplex, and T-A-T Triplex Structures, Adv. Funct. Mater., 2023, 33, 2304966 Search PubMed.
- Y. Xing, E. Cheng, Y. Yang, P. Chen, T. Zhang, Y. Sun, Z. Yang and D. Liu, Self-assembled DNA hydrogels with designable thermal and enzymatic responsiveness, Adv. Mater., 2011, 23, 1117–1121 CrossRef CAS PubMed.
- Y. Si, L. Xu, N. Wang, J. Zheng, R. Yang and J. Li, Target microRNA-responsive DNA hydrogel-based surface-enhanced Raman scattering sensor arrays for microRNA-marked cancer screening, Anal. Chem., 2020, 92, 2649–2655 Search PubMed.
- A. Fischer, P. Zhang, Y. Ouyang, Y. S. Sohn, O. Karmi, R. Nechushtai, E. Pikarsky and I. Willner, DNA-Tetrahedra Corona-Modified Hydrogel Microcapsules:“Smart” ATP-or microRNA-Responsive Drug Carriers, Small, 2022, 18, 2204108 CrossRef CAS PubMed.
- M. H. Lee, Z. Yang, C. W. Lim, Y. H. Lee, S. Dongbang, C. Kang and J. S. Kim, Disulfide-cleavage-triggered chemosensors and their biological applications, Chem. Rev., 2013, 113, 5071–5109 Search PubMed.
- S. Liu, W. Su, Y. Li, L. Zhang and X. Ding, Manufacturing of an electrochemical biosensing platform based on hybrid DNA hydrogel: Taking lung cancer-specific miR-21 as an example, Biosens. Bioelectron., 2018, 103, 1–5 CrossRef CAS PubMed.
- Y. Zhang, W. Wang, X. Zhou, H. Lin, X. Zhu, Y. Lou and L. Zheng, CRISPR-responsive RCA-based DNA hydrogel biosensing platform with customizable signal output for rapid and sensitive nucleic acid detection, Anal. Chem., 2024, 96, 15998–16006 Search PubMed.
- E. Ahmadi-Sangachin, J. Mohammadnejad and M. Hosseini, Fluorescence self-assembled DNA hydrogel for the determination of prostate specific antigen by aggregation induced emission, Spectrochim. Acta, Part A, 2023, 303, 123234 CrossRef CAS PubMed.
- C.-H. Lu, W. Guo, Y. Hu, X.-J. Qi and I. Willner, Multitriggered shape-memory acrylamide–DNA hydrogels, J. Am. Chem. Soc., 2015, 137, 15723–15731 CrossRef CAS PubMed.
- W. Guo, C. H. Lu, X. J. Qi, R. Orbach, M. Fadeev, H. H. Yang and I. Willner, Switchable bifunctional stimuli-triggered poly-N-isopropylacrylamide/DNA hydrogels, Angew. Chem., Int. Ed., 2014, 53, 10134–10138 CrossRef CAS PubMed.
- Q. Chen, S. Wang, T. Huang, F. Xiao, Z. Wu and R. Yu, Construction and Research of Multiple Stimuli-Responsive 2D Photonic Crystal DNA Hydrogel Sensing Platform with Double-Network Structure and Signal Self-Expression, Anal. Chem., 2022, 94(14), 5530–5537 CrossRef CAS PubMed.
- C. Wang, F. Li, Y. Bi and W. Guo, Reversible Modulation of 2D Photonic Crystals with a Responsive Shape-Memory DNA Hydrogel Film, Adv. Mater. Interfaces, 2019, 6, 1900556 CrossRef CAS.
- A. Cangialosi, C. Yoon, J. Liu, Q. Huang, J. Guo, T. D. Nguyen, D. H. Gracias and R. Schulman, DNA sequence–directed shape change of photopatterned hydrogels via high-degree swelling, Science, 2017, 357, 1126–1130 CrossRef CAS PubMed.
- Y. Zhang, L. Zhu, J. Tian, L. Zhu, X. Ma, X. He, K. Huang, F. Ren and W. Xu, Smart and functionalized development of nucleic acid-based hydrogels: Assembly strategies, recent advances, and challenges, Adv. Sci., 2021, 8, 2100216 CrossRef CAS PubMed.
- J. Gačanin, C. V. Synatschke and T. Weil, Biomedical applications of DNA-based hydrogels, Adv. Funct. Mater., 2020, 30, 1906253 CrossRef.
- I. Zare, R. Taheri-Ledari, F. Esmailzadeh, M. M. Salehi, A. Mohammadi, A. Maleki and E. Mostafavi, DNA hydrogels and nanogels for diagnostics, therapeutics, and theragnostics of various cancers, Nanoscale, 2023, 15, 10882–10903 RSC.
- A. Chakraborty, S. P. Ravi, Y. Shamiya, C. Cui and A. Paul, Harnessing the physicochemical properties of DNA as a multifunctional biomaterial for biomedical and other applications, Chem. Soc. Rev., 2021, 50, 7779–7819 RSC.
- M. W. Tibbitt, J. E. Dahlman and R. Langer, Emerging frontiers in drug delivery, J. Am. Chem. Soc., 2016, 138, 704–717 CrossRef CAS PubMed.
- L. Zhou, X. Jiao, S. Liu, M. Hao, S. Cheng, P. Zhang and Y. Wen, Functional DNA-based hydrogel intelligent materials for biomedical applications, J. Mater. Chem. B, 2020, 8, 1991–2009 RSC.
- T. Kim, S. Park, M. Lee, S. Baek, J. B. Lee and N. Park, DNA hydrogel microspheres and their potential applications for protein delivery and live cell monitoring, Biomicrofluidics, 2016, 10, 034112 CrossRef PubMed.
- F. Li, J. Tang, J. Geng, D. Luo and D. Yang, Polymeric DNA hydrogel: Design, synthesis and applications, Prog. Polym. Sci., 2019, 98, 101163 CrossRef CAS.
- Y. Hu, S. Gao, H. Lu and J. Y. Ying, Acid-resistant and physiological pH-responsive DNA hydrogel composed of A-motif and i-motif toward oral insulin delivery, J. Am. Chem. Soc., 2022, 144, 5461–5470 CrossRef CAS PubMed.
- X. Fu, T. Chen, Y. Song, C. Feng, H. Chen, Q. Zhang, G. Chen and X. Zhu, mRNA Delivery by a pH-Responsive DNA Nano-Hydrogel, Small, 2021, 17, 2101224 Search PubMed.
- C. Li, Y. Li, B. Zhou, T. Li, X. Wei, K. Chen, W. Chen, Z. Shi, X. Dai and J. Zhang, Enantiomer-dependent and modification-free DNA matrix as an adjuvant for subunit vaccines against SARS-CoV-2 or pneumococcal infections, Nat. Biomed. Eng., 2025, 1–25 Search PubMed.
- Y. Shao, Z.-Y. Sun, Y. Wang, B.-D. Zhang, D. Liu and Y.-M. Li, Designable immune therapeutical vaccine system based on DNA supramolecular hydrogels, ACS Appl. Mater. Interfaces, 2018, 10, 9310–9314 Search PubMed.
- D. Wang, J. Liu, J. Duan, H. Yi, J. Liu, H. Song, Z. Zhang, J. Shi and K. Zhang, Enrichment and sensing tumor cells by embedded immunomodulatory DNA hydrogel to inhibit postoperative tumor recurrence, Nat. Commun., 2023, 14, 4511 Search PubMed.
- Y. Fan, M. Zhan, J. Liang, X. Yang, B. Zhang, X. Shi and Y. Hu, Programming Injectable DNA Hydrogels Yields Tumor Microenvironment-Activatable and Immune-Instructive Depots for Augmented Chemo-Immunotherapy, Adv. Sci., 2023, 10, 2302119 Search PubMed.
- I. Mellman, G. Coukos and G. Dranoff, Cancer immunotherapy comes of age, Nature, 2011, 480, 480–489 Search PubMed.
- Q. Liu, J. Li, H. Zheng, S. Yang, Y. Hua, N. Huang, J. Kleeff, Q. Liao and W. Wu, Adoptive cellular immunotherapy for solid neoplasms beyond CAR-T, Mol. Cancer, 2023, 22, 28 CrossRef PubMed.
- A. Ribas and J. D. Wolchok, Cancer immunotherapy using checkpoint blockade, Science, 2018, 359, 1350–1355 CrossRef CAS PubMed.
- Q. Chen, Y. Liu, Q. Chen, M. Li, L. Xu, B. Lin, Y. Tan and Z. Liu, DNA nanostructures: Advancing cancer immunotherapy, Small, 2024, 20, 2405231 Search PubMed.
- J. Sun, M. Pan, F. Liu, W. Yu, W. Wang, F. Mo, S. Yu, Y. Zhou and X. Liu, Immunostimulatory DNA nanogel enables effective lymphatic drainage and high vaccine efficacy, ACS Mater. Lett., 2020, 2, 1606–1614 CrossRef CAS.
- T. Yata, Y. Takahashi, M. Tan, H. Nakatsuji, S. Ohtsuki, T. Murakami, H. Imahori, Y. Umeki, T. Shiomi and Y. Takakura, DNA nanotechnology-based composite-type gold nanoparticle-immunostimulatory DNA hydrogel for tumor photothermal immunotherapy, Biomaterials, 2017, 146, 136–145 CrossRef CAS PubMed.
- C. Frantz, K. M. Stewart and V. M. Weaver, The extracellular matrix at a glance, J. Cell Sci., 2010, 123, 4195–4200 Search PubMed.
- H. K. Kleinman, D. Philp and M. P. Hoffman, Role of the extracellular matrix in morphogenesis, Curr. Opin. Biotechnol., 2003, 14, 526–532 CrossRef CAS PubMed.
- S. R. Caliari and J. A. Burdick, A practical guide to hydrogels for cell culture, Nat. Methods, 2016, 13, 405–414 CrossRef CAS PubMed.
- J. Lou and D. J. Mooney, Chemical strategies to engineer hydrogels for cell culture, Nat. Rev. Chem., 2022, 6, 726–744 CrossRef CAS PubMed.
- M. J. Webber, E. A. Appel, E. W. Meijer and R. Langer, Supramolecular biomaterials, Nat. Mater., 2016, 15, 13–26 CrossRef CAS PubMed.
- J. L. Mann, A. C. Yu, G. Agmon and E. A. Appel, Supramolecular polymeric biomaterials, Biomater. Sci., 2018, 6, 10–37 RSC.
- G. Li, F. Gao, D. Yang, L. Lin, W. Yu, J. Tang, R. Yang, M. Jin, Y. Gu and P. Wang, ECM-mimicking composite hydrogel for accelerated vascularized bone regeneration, Bioact. Mater., 2024, 42, 241–256 CAS.
- M. Zhu, H. Zhang, Q. Zhou, S. Sheng, Q. Gao, Z. Geng, X. Chen, Y. Lai, Y. Jing, K. Xu, L. Bai, G. Wang, J. Wang, Y. Jiang and J. Su, Dynamic GelMA/DNA Dual-Network Hydrogels Promote Woven Bone Organoid Formation and Enhance Bone Regeneration, Adv. Mater., 2025, 37, 2501254 Search PubMed.
- K. Nam, B. I. Im, T. Kim, Y. M. Kim and Y. H. Roh, Anisotropically Functionalized Aptamer-DNA Nanostructures for Enhanced Cell Proliferation and Target-Specific Adhesion in 3D Cell Cultures, Biomacromolecules, 2021, 22, 3138–3147 CrossRef CAS PubMed.
- J. Tang, J. Wang, J. Ou, Z. Cui, C. Yao and D. Yang, A DNA/Poly-(L-lysine) Hydrogel with Long Shelf-Time for 3D Cell Culture, Small Methods, 2024, 8, 2301236 CrossRef CAS PubMed.
- B. Yang, B. Zhou, C. Li, X. Li, Z. Shi, Y. Li, C. Zhu, X. Li, Y. Hua, Y. Pan, J. He, T. Cao, Y. Sun, W. Liu, M. Ge, Y. R. Yang, Y. Dong and D. Liu, A Biostable l-DNA Hydrogel with Improved Stability for Biomedical Applications, Angew. Chem., Int. Ed., 2022, 61, e202202520 Search PubMed.
- J. Wu, B. R. Liyarita, H. Zhu, M. Liu, X. Hu and F. Shao, Self-Assembly of Dendritic DNA into a Hydrogel: Application in Three-Dimensional Cell Culture, ACS Appl. Mater. Interfaces, 2021, 13, 49705–49712 Search PubMed.
- Y. Mu, X. Wang, X. Du, P.-P. He and W. Guo, DNA Cryogels with Anisotropic Mechanical and Responsive Properties for Specific Cell Capture and Release, J. Am. Chem. Soc., 2024, 146, 5998–6005 Search PubMed.
- Y. Miao, X. Liu, J. Luo, Q. Yang, Y. Chen and Y. Wang, Double-Network DNA Macroporous Hydrogel Enables Aptamer-Directed Cell Recruitment to Accelerate Bone Healing, Adv. Sci., 2024, 11, 2303637 CrossRef CAS PubMed.
- C. Yao, H. Tang, W. Wu, J. Tang, W. Guo, D. Luo and D. Yang, Double Rolling Circle Amplification Generates Physically Cross-Linked DNA Network for Stem Cell Fishing, J. Am. Chem. Soc., 2020, 142, 3422–3429 Search PubMed.
- P. Song, D. Ye, X. Zuo, J. Li, J. Wang, H. Liu, M. T. Hwang, J. Chao, S. Su, L. Wang, J. Shi, L. Wang, W. Huang, R. Lal and C. Fan, DNA Hydrogel with Aptamer-Toehold-Based Recognition, Cloaking, and Decloaking of Circulating Tumor Cells for Live Cell Analysis, Nano Lett., 2017, 17, 5193–5198 Search PubMed.
- R. Zhang, Z. Lv, L. Chang, J. Wang, J. Tang, Z. Wang, S. Li, J. Guo, C. Yao and D. Yang, A Responsive DNA Hydrogel Containing Poly-Aptamers as Dual-Target Inhibitors for Localized Cancer Immunotherapy, Adv. Funct. Mater., 2024, 34, 2401563 CrossRef CAS.
- Z. Zhu, Y. Yang, Y. Jiang, T. Gu, L. Siow, Y. Gao, Y. Zheng, K. Xing, S. Zhou and C. Zhang, DNA Hydrogels in Tissue Engineering: From Molecular Design to Next-Generation Biomedical Applications, Adv. Healthcare Mater., 2025, 14, 2500192 Search PubMed.
- R. Wu, W. Li, P. Yang, N. Shen, A. Yang, X. Liu, Y. Ju, L. Lei and B. Fang, DNA hydrogels and their derivatives in biomedical engineering applications, J. Nanobiotechnol., 2024, 22, 518 Search PubMed.
- T. Yuan, Y. Shao, X. Zhou, Q. Liu, Z. Zhu, B. Zhou, Y. Dong, N. Stephanopoulos, S. Gui and H. Yan, Highly permeable DNA supramolecular hydrogel promotes neurogenesis and functional recovery after completely transected spinal cord injury, Adv. Mater., 2021, 33, 2102428 Search PubMed.
- C. Shen, J. Wang, G. Li, S. Hao, Y. Wu, P. Song, Y. Han, M. Li, G. Wang and K. Xu, Boosting cartilage repair with silk fibroin-DNA hydrogel-based cartilage organoid precursor, Bioact. Mater., 2024, 35, 429–444 Search PubMed.
- R. Ye, Z. Zhu, T. Gu, D. Cao, K. Jiang, Q. Dai, K. Xing, Y. Jiang, S. Zhou and P. Cai, Neutrophil extracellular traps-inspired DNA hydrogel for wound hemostatic adjuvant, Nat. Commun., 2024, 15, 5557 CrossRef CAS PubMed.
- X. Yan, B. Yang, Y. Chen, Y. Song, J. Ye, Y. Pan, B. Zhou, Y. Wang, F. Mao and Y. Dong, Anti-friction MSCs delivery system improves the therapy for severe osteoarthritis, Adv. Mater., 2021, 33, 2104758 CrossRef CAS PubMed.
- M. Xiong, X. Yang, Z. Shi, J. Xiang, H. Gao, S. Ji, Y. Li, W. Pi, H. Chen and H. Zhang, Programmable artificial skins accomplish antiscar healing with multiple appendage regeneration, Adv. Mater., 2024, 36, 2407322 Search PubMed.
- L. Zhou, W. Pi, S. Cheng, Z. Gu, K. Zhang, T. Min, W. Zhang, H. Du, P. Zhang and Y. Wen, Multifunctional DNA hydrogels with hydrocolloid-cotton structure for regeneration of diabetic infectious wounds, Adv. Funct. Mater., 2021, 31, 2106167 CrossRef CAS.
- Y. Jin, Y. Li, S. Song, Y. Ding, Y. Dong, Y. Lu, D. Liu and C. Zhang, DNA supramolecular hydrogel as a biocompatible artificial vitreous substitute, Adv. Mater. Interfaces, 2022, 9, 2101321 Search PubMed.
- P. Bannigan, Z. Bao, R. J. Hickman, M. Aldeghi, F. Häse, A. Aspuru-Guzik and C. Allen, Machine learning models to accelerate the design of polymeric long-acting injectables, Nat. Commun., 2023, 14, 35 Search PubMed.
- F. Li, J. Han, T. Cao, W. Lam, B. Fan, W. Tang, S. Chen, K. L. Fok and L. Li, Design of self-assembly dipeptide hydrogels and machine learning via their chemical features, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 11259–11264 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2026 |





