Open Access Article
Open Access ArticleA printable, unimolecular, core–shell polymer bottlebrush-based signal transducer using solvatochromatic reporting
Chenyou
Zhang
a,
Samantha O.
Catt
 b,
Tom
Hawtrey
cde,
Ping
Zeng
a,
Haoxiang
Zeng
b,
Tom
Hawtrey
cde,
Ping
Zeng
a,
Haoxiang
Zeng
 a,
Simran D.
Kerai
a,
Simran D.
Kerai
 a,
Yen Theng
Cheng
a,
Moritz P.
Hopp
b,
Qinqin
Yang
a,
Elizabeth J.
New
a,
Yen Theng
Cheng
a,
Moritz P.
Hopp
b,
Qinqin
Yang
a,
Elizabeth J.
New
 cde,
Eva
Blasco
cde,
Eva
Blasco
 *b and
Markus
Müllner
*b and
Markus
Müllner
 *ad
*ad
aKey Centre for Polymers & Colloids, School of Chemistry, The University of Sydney, Sydney, NSW 2006, Australia. E-mail: markus.muellner@sydney.edu.au
bInstitute for Molecular Systems Engineering and Advanced Materials (IMSEAM), Heidelberg University, 69120 Heidelberg, Germany. E-mail: eva.blasco@uni-heidelberg.de
cSchool of Chemistry, The University of Sydney, Sydney, NSW 2006, Australia
dThe University of Sydney Nano Institute (Sydney Nano), The University of Sydney, Sydney, NSW 2006, Australia
eAustralian Research Council Centre of Excellence for Innovations in Peptide and Protein Science, The University of Sydney, Sydney, 2006 NSW, Australia
First published on 18th November 2025
Abstract
The combination of stimuli-responsiveness, colloidal stability and tolerance to changes in concentration and ionic strength into a functional nanoparticle system is challenging. Self-assembled polymer materials remain susceptible to environmental changes, which in turn limit their application potential. Unimolecular architectures like bottlebrush polymers offer enhanced stability and functional compartmentalisation within a single macromolecule. Here, we report the synthesis of a pH-responsive molecular polymer bottlebrush (MPB) featuring a poly[(diisopropylamino)ethyl methacrylate] (PDPAEMA) core, a hydrophilic poly[poly(ethylene glycol) methyl ether methacrylate] (PPEGMA) shell, and naphthalimide-based solvatochromic fluorophores embedded within its core. The PDPAEMA segment imparts pH sensitivity, while the PEGMA shell improves colloidal stability and biocompatibility. The solvatochromic dyes report environmental changes by responding to polarity shifts in the polymer architecture and polymer networks. Our proof-of-concept study promises future application of such unimolecular constructs as nanoscale pH sensors for biomedicine or as functional additives for 3D printing capable of reporting environmental changes within a printed network or (hydro)gel.
Introduction
The architectural design of macromolecular systems fundamentally determines their functional capabilities and potential applications.1,2 Macromolecules can exist as unimolecular entities or as multi-molecular assemblies, with each paradigm offering distinct advantages and limitations. Amphiphilic block copolymers self-assemble into micellar structures once a critical unimer concentration is reached. These self-assembled materials have found widespread applications from health to energy.3–6 Importantly, properties can be readily tuned as the segregation of the micelle core and shell offer distinct microenvironments that can be separately addressed. However, conventional polymeric micelles are thermodynamic aggregates that suffer from inherent instability when subjected to surrounding environment changes such as high dilution and alterations in temperature, pH, and ionic strength, often leading to disassembly into free polymeric chains.7In recent years, unimolecular polymer particle systems, like star and bottlebrush polymers, have emerged as a compelling alternative to overcome these issues associated with thermodynamic instability.8–11 These covalent nanomaterials allow for the generation of core–shell microenvironments within one macromolecule, ensuring stability while also enabling compartmentalisation of function.12,13 This unique architectural improvement has made them particularly attractive in specific applications, especially in nanomedicine and template chemistry, where the use of such polymer topologies has risen sharply in recent years. Molecular polymer bottlebrushes (MPBs, or bottlebrush polymers) in particular have become a versatile class of materials which is emerging from a niche in fundamental research on their synthesis into application-driven research on therapeutics, theranostics, additives, 3D printing and photonics.14–20
One important application of polymeric systems is the detection of environmental changes, such as pH, temperature, or ionic strength.21 Traditional approaches involved decorating polymers with small molecule fluorophores which themselves can detect the change.22 However, the use of fluorophores that are directly sensitive to the change, e.g. pH-dependent protonation/deprotonation, can lead to complications in signal interpretation due to on/off responses or loss of fluorescent properties. An alternative approach involves the design of polymers that are themselves responsive to pH, changing their properties whether by dehydration, conformational transitions, or aggregation. Such polymers can then be coupled to fluorophores that are insensitive to pH but are sensitive to their microenvironment. Ideal candidates for this approach are solvatochromic dyes,23 whose emission wavelength depends on the polarity of their microenvironment.24,25 When incorporated into pH-responsive unimolecular polymers, these fluorophores may provide optical readouts of environmental changes through polymer matrix-mediated alterations in local polarity. The potential for bottlebrush polymers in this context has not been explored, despite the opportunity to design nanoparticles with controllable endocytosis for nanomedicine applications,26,27 where extracting information on intracellular pH remains challenging to determine. Similarly, MPBs can be used in 3D printing applications,15 whereby a functional ink that can report environmental changes within the print would offer new insight into local microenvironments.
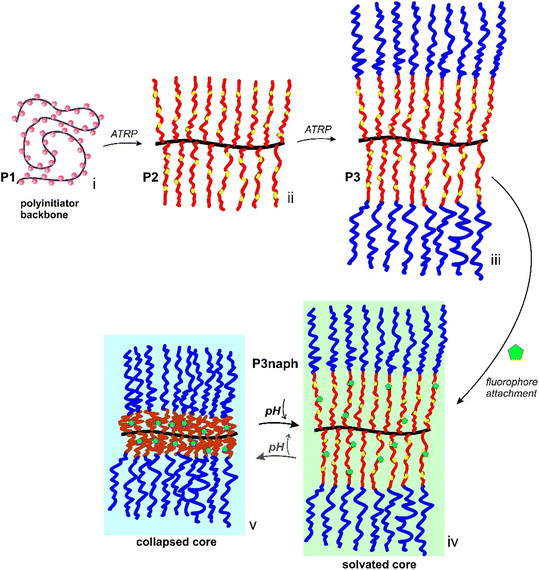
Herein we established a proof of concept toward unimolecular signal transducers for pH measurements. We describe the synthesis of a unimolecular core–shell MPB consisting of a pH-responsive poly[(diisopropylamino)ethyl methacrylate] (PDPAEMA) core, a poly[poly(ethylene glycol) methyl ether methacrylate] (PPEGMA) shell and naphthalimide-based fluorophores within the core (Scheme 1).
Each synthetic step is described in detail in the Experimental section (SI). To yield our core–shell MPB, we first prepared a poly(methyl methacrylate-co-2-(2-bromoisobutyryloxy)ethyl methacrylate) (PMMA25-co-PBIEM25) polyinitiator (P1). As grafting efficiency from pure PBIEM is only between 50–70%,28–30 we opted for a copolymer backbone which is easier to handle without compromising the MPB synthesis. Subsequently, we grafted a core consisting of a copolymer of (diisopropylamino)ethyl methacrylate (DPAEMA) and tert-butyloxycarbonyl (boc)-protected aminoethyl methacrylate (bocAEMA) (ratio 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
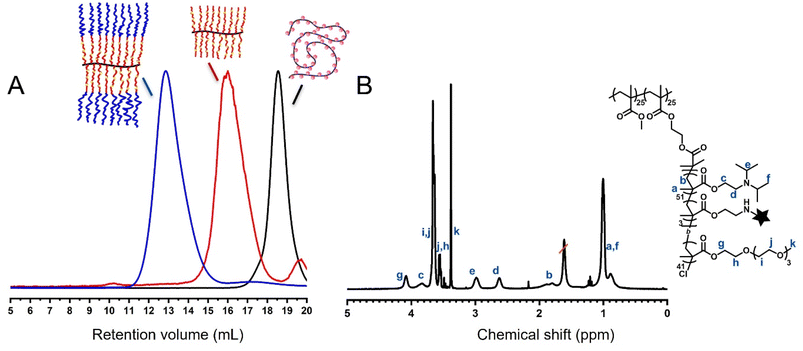
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 mol%) to yield P2. PDPAEMA is a well-known pH-responsive polymer with a pKa of ∼6.3–6.8,31,32 while the incorporated bocAEMA units serve as latent amines for post-functionalisation within the core. The relatively low concentration of dye attachment sites is to minimise fluorophore–fluorophore interactions and to avoid changes to the responsiveness of the core which may interfere with fluorescence intensity and interpretation. Next, a hydrophilic, charge-neutral shell of PEGMA was grafted from P2 through chain extension yielding the core–shell MPB P3. The purpose of the shell is to increase colloidal stability and shield the core from intermolecular interactions.33 Removal of the boc group then allowed for the selective attachment of naphthalimide fluorophores (naphmal) to the core to yield our unimolecular signal transducer P3naph. The 4-amino-1,8-naphthalimide derived fluorophore was chosen as it is known to display solvatochromic behaviour due to the effect of solvent interactions on its intramolecular charge transfer (ICT) character.34,35 This property has allowed related naphthalimides to be used to study dynamic protein interactions,36 in assays detecting ct-DNA,37 and for distinguishing different organelles in cell imaging.38,39 Naphthalimides can be designed to act as pH-responsive fluorophores themselves; however, this requires additional synthetic steps, for example, the introduction of functionalities to facilitate photoinduced electron transfer (PET) quenching.40 The stepwise build-up of the core–shell MPBs was followed by size exclusion chromatography (SEC) (Fig. 1A), with the significant shifts in elution volumes after each grafting step indicating a steady growth in apparent hydrodynamic volume (i.e. molecular weight increase), along with relatively low dispersity. Fig. 1B shows a representative proton NMR spectrum of the final core–shell MPB. Complete 1H NMR spectra for P0, P1, P2 and P3 are provided in the Experimental section (SI, Fig. S1–S4). Table 1 summarises key parameters of the polymerisations and MPBs.
5 mol%) to yield P2. PDPAEMA is a well-known pH-responsive polymer with a pKa of ∼6.3–6.8,31,32 while the incorporated bocAEMA units serve as latent amines for post-functionalisation within the core. The relatively low concentration of dye attachment sites is to minimise fluorophore–fluorophore interactions and to avoid changes to the responsiveness of the core which may interfere with fluorescence intensity and interpretation. Next, a hydrophilic, charge-neutral shell of PEGMA was grafted from P2 through chain extension yielding the core–shell MPB P3. The purpose of the shell is to increase colloidal stability and shield the core from intermolecular interactions.33 Removal of the boc group then allowed for the selective attachment of naphthalimide fluorophores (naphmal) to the core to yield our unimolecular signal transducer P3naph. The 4-amino-1,8-naphthalimide derived fluorophore was chosen as it is known to display solvatochromic behaviour due to the effect of solvent interactions on its intramolecular charge transfer (ICT) character.34,35 This property has allowed related naphthalimides to be used to study dynamic protein interactions,36 in assays detecting ct-DNA,37 and for distinguishing different organelles in cell imaging.38,39 Naphthalimides can be designed to act as pH-responsive fluorophores themselves; however, this requires additional synthetic steps, for example, the introduction of functionalities to facilitate photoinduced electron transfer (PET) quenching.40 The stepwise build-up of the core–shell MPBs was followed by size exclusion chromatography (SEC) (Fig. 1A), with the significant shifts in elution volumes after each grafting step indicating a steady growth in apparent hydrodynamic volume (i.e. molecular weight increase), along with relatively low dispersity. Fig. 1B shows a representative proton NMR spectrum of the final core–shell MPB. Complete 1H NMR spectra for P0, P1, P2 and P3 are provided in the Experimental section (SI, Fig. S1–S4). Table 1 summarises key parameters of the polymerisations and MPBs.
| Composition | Conversiona (%) | Feed ratiob | M n,NMR (kg mol−1) | M n,SEC (kg mol−1) | Đ | Hydrodynamic diameterd (nm) | |
|---|---|---|---|---|---|---|---|
a Conversion was determined by 1H NMR spectroscopy. Mn,NMR was calculated considering 100% grafting efficiency from P1.
b [Polyinitiator]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [monomer] [monomer]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [ligand] [ligand]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [copper halide] (halide) molar feed ratio for the ATRP.
c Apparent average molecular weight Mn,SEC obtained from DMAc SEC using monodisperse PMMA calibration standards.
d
Z-Average of DLS measurements of polymers in chloroform-d. [copper halide] (halide) molar feed ratio for the ATRP.
c Apparent average molecular weight Mn,SEC obtained from DMAc SEC using monodisperse PMMA calibration standards.
d
Z-Average of DLS measurements of polymers in chloroform-d.
|
|||||||
| P0 | PMMA25-co-PHEMA25 | 99 | [1]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [25/25] [25/25]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [1.5] [1.5]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [1.1] (Cl) [1.1] (Cl) |
10.3 | 12 | 1.16 | — |
| P1 | PMMA25-co-PBIEM25 | — | — | 9.7 | 10.1 | 1.18 | — |
| P2 | [DPAEMA51-co-bocAEMA3] | 24 | [1]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [190/10] [190/10]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2] [2]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [1.2] (Br) [1.2] (Br) |
45.4 | 51.1 | 1.13 | 19 |
| P3 | [(DPAEMA51-co-bocAEMA3)-b-PPEGMA41] | 23 | [1]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [200] [200]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [4] [4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2] (Cl) [2] (Cl) |
215 | 262 | 1.22 | 34 |
We initially tested the deprotection and fluorophore conjugation steps as an effective post-functionalisation strategy on a linear copolymer analogue poly(DPAEMA30-co-bocAEMA3) (SI, Fig. S5). 1H NMR spectroscopy revealed the successful removal of boc groups through the disappearance of proton signals at 1.44 ppm, while successful fluorophore conjugation was evidenced by the appearance of proton signals at around ∼7 ppm, corresponding to the aromatic protons of naphthalimide (Fig. S6). For the MPB, where conjugation occurs within the shielded core, successful conjugation of the naphthalimide fluorophore was confirmed qualitatively through the fluorescence measurements of P3naph, which closely matched that of the free naphmal (Fig. S7A). In addition, the yellow colouration of P3naph solution after extensive dialysis further supports successful incorporation of the naphthalimide fluorophore (Fig. S7B).
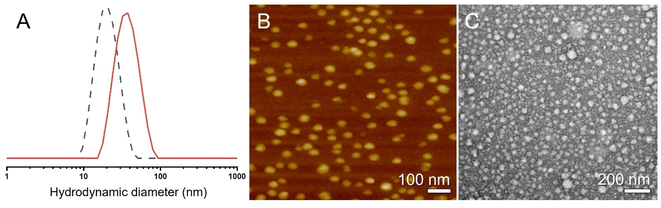
Next, we used dynamic light scattering (DLS), atomic force microscopy (AFM), and transmission electron microscopy (TEM) to determine the size of the unimolecular MPBs (Fig. 2). The hydrodynamic diameter of P3 in deuterated chloroform was around 34 nm (Fig. 2A, red). While AFM and TEM measurements (Fig. 2B and C) are conducted on substrates in the dry state, they did also reveal a unimolecular character with comparable nanoscale dimensions. DLS further demonstrated the expected increase in hydrodynamic diameter from P2 (19 nm) to P3 (34 nm).
As the PDPAEMA core is pH-responsive, it is expected that the core polymer alone will precipitate at neutral or higher pH and only be soluble if the pH is sufficiently low to guarantee hydrophilicity of the protonated DPAEMA units.41–43 Given the core–shell character of our MPBs, the pH-responsiveness is contained within the core and its collapse can only occur intramolecularly without affecting the unimolecular characteristics of the MPBs (Fig. 3A and B). The responsiveness of the core was evident in DLS where the hydrodynamic diameter of the MPBs was the smallest at neutral pH (i.e. deprotonated, hydrophobic core) and slightly increased towards acidic pH (i.e. protonated, hydrophilic core) (Fig. 3B). This switchability of the core provides a polarity-tuneable internal environment within the core–shell MPB nanoparticle. Since the solvatochromic naphthalimide fluorophore resides within the core, the fluorophore experiences the (de)hydration of the core environment upon pH changes and alters its fluorescence. The shift in core polarity translates into a modulated fluorescence output, underlining that local environment changes are effectively transduced through the fluorophore. Photophysical studies on the free naphmal affirm this behaviour and demonstrate insensitivity to pH (Fig. S8 and S9). Below the pKa of PDPAEMA, protonation of the tertiary amines bestows hydrophilicity, facilitating a polar core environment. At a pH above its pKa, deprotonation renders the core more hydrophobic, activating an apolar microenvironment. Thus, fluorescence emission spectra were obtained in buffer solutions of P3naph at pH values above and below its pKa (Fig. 3C and S10).
The solvatochromic nature of naphthalimides is thoroughly studied in the literature.44,45 Our naphthalimide fluorophore itself does not show a pH response (Fig. S9), thus corroborating that the fluorescence changes in our P3naph stem from a change in their core environments which was caused by the pH change. This distinction between traditional small molecule sensors, whose sensitivity is afforded by their intrinsic chemical and electronic structure,46 and the presented MPB platform is noteworthy. We describe here a signal transduction platform, where solvatochromic fluorophores can be ‘sensitised’ to a stimulus (e.g. pH), which they would otherwise be insensitive to.
The onset of the fluorescence red shift in P3naph occurred at pH below the nominal pKa of PDPAEMA, with fluorescent and size changes observed near pH 5.8 (Fig. S10 and S11). This indicates a more complex relationship between pH and the core hydrophobicity than can be explained by simple protonation equilibrium alone. Potential contributing factors include stabilising hydrophobic interactions among the diisopropyl groups,41,47 exacerbated by their proximity within a polymer brush,47 and partial (de)protonation across the polymer chains. These contributions find some further proof in diluting the buffers used, i.e. on reducing ionic strength, at pH = 5.1 below the switching pH = 5.8 noted earlier, the MPB system can be actuated (Fig. S12). This demonstration of the ion screening effect exposes additional complex intersystem interactions beyond the previously thought model. More detailed work is necessary to fine-tune the response of the fluorophore, for example via a different core material or fluorophore class to develop this concept further to allow a more linear response to pH changes. Our current observations however highlight the opportunity of nanoscale architecture design to utilise stimulus-responsiveness within unimolecular nanoparticles. Nevertheless, the change in fluorescence affirms the functionality of the P3naph signal transducer platform, which can be further optimised and explored for specific applications. As previously mentioned, the use of MPBs in the biomedical context has grown enormously in the last decade, and herein we have also verified that the current unimolecular system is compatible with cells and are readily taken up into cancer cells (Fig. S13 and S14). Furthermore, the unimolecular character offers another opportunity to impart new properties to gels/prints, which has not been explored to the best of our knowledge.
Two-photon laser printing (2PLP) has become a powerful fabrication tool to produce nano- and micro-structured materials with high precision. Particularly, polymer-based inks are heavily investigated as they endow the printed material with excellent versatility and adaptability. MPBs offer an unexplored opportunity to design functional inks for 3D printing applications. Thus, formulating tuneable inks with the capability to produce stable, crosslinked networks while at the same time having stimuli-responsiveness incorporated, is desirable. To test whether our MPBs can be printed with 2PLP while retaining their functionality to transduce pH changes, we redesigned our MPBs and installed methacrylate functionality as a photoactive crosslinkable group onto its shell.48,49 We achieved this by grafting a copolymer of PEGMA-OMe and PEGMA-OH as the shell. This enabled the installation of methacrylate moieties onto PEGMA-OH within the shell while leaving the core functionality unchanged (Fig. S15). Subsequently, the photoactive MPB was formulated into a suitable ink composition for 2PLP. The synthesis protocol and ink formulation can be found in the SI. Using a two-photon laser printer (PPGT2, Nanoscribe GmbH), a snowflake pattern was printed (Fig. 4), and the resultant structures were swollen in aqueous media. In this way, we were able to incorporate fluorescent MPBs into high fidelity microscale prints (Fig. 4A and B). In addition, not only were our printed materials able to incorporate the fluorescence of naphthalimide, but more importantly, the responsiveness to changes in pH was carried over into the final structures (Fig. 4C). Incubation of the hydrogels at different pH values resulted in fluorescence changes which – like our unimolecular system – stems from the (de)hydration of the PDPAEMA core within the MPBs that make up the print. Spectral scanning using a Stellaris Falcon confocal microscope reported the expected spectral shift within the prints, as observed before in the unimolecular MPBs in aqueous media (Fig. S16).
Conclusions
We presented a colloidally stable unimolecular core–shell nanomaterial capable of responding to pH changes by altering its fluorescence profile. Specifically, our MPB system exploits a pH sensitive core to alter the core microenvironment to sensitise a co-localised solvatochromic naphthalimide dye. This pH-dependent fluorescence change could further be carried over into a printed hydrogel microstructure obtained using two-photon polymerisation. Our proof-of-concept study sets the groundwork for advanced sensors that may become useful for additive manufacturing, such as inks for hydrogels that can simultaneously report on environmental changes, or in biomedicine, where a PEGylated nanoparticle can enter cells or tumour environments to read out environmental changes as it is processed by the body.Author contributions
MM conceived the idea. CZ and MM conceptualised the project. CZ designed, synthesised, characterised, and analysed the polymers, and their modifications, responses, and printed products. SOC and CZ performed modifications for 2PLP. SOC and MPH performed 2PLP printing and characterisation under supervision from EB. TH synthesised and characterised the fluorophore, under supervision from EJN. PZ and QY performed confocal microscopy studies. PZ carried out cell studies and analysed the results. HZ performed TEM. SDK performed AFM. YTC performed SEM. CZ and MM co-wrote the manuscript, with input from all authors.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc07029e.Acknowledgements
This project was supported by the UA-DAAD Australia–Germany Joint Research Cooperation Scheme, an initiative of Universities Australia (UA) and the German Academic Exchange Service (DAAD). Our research was facilitated by access to Sydney Analytical, a core research facility at the University of Sydney. The authors acknowledge the technical and scientific assistance from Sydney Microscopy & Microanalysis, the University of Sydney node of Microscopy Australia. C. Z. is grateful to Finn Kröger (Heidelberg University) for 2PLP and fluorescence microscopy investigations and valuable discussions; and Dr Pamela Young (Sydney Microscopy & Microanalysis) for spectral imaging confocal microscopy. The acquisition of the microscope was funded through an ARC LIEF grant (LE240100091). We thank Professor Ronald Clarke for access to a fluorometer, Professor Girish Lakhwani and Pratiksha Dad for valuable discussions, and Professor Chiara Neto for access to an atomic force microscope. C. Z. gratefully acknowledges the L. E. R. Tonnet and Family Scholarship in Chemistry, and a supplementary scholarship from the School of Chemistry, University of Sydney. P. Z. was supported by the Postgraduate Research Scholarships scheme from The University of Sydney. S. D. K. is a recipient of an Australian Government RTP scholarship. Q. Y. is a recipient of the Henry Bertie and Florence Mabel Gritton Research Scholarship. M. M. acknowledges the Australian Research Council for a Future Fellowship (FT200100185) and Discovery Project (DP220100452), respectively. M. M. is a grateful recipient of a Global Science and Technology Diplomacy Fund (GSTDS000001 – 6) grant. SOC, MPH, and EB acknowledge the funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) via the Excellence Cluster “3D Matter Made to Order” (EXC-2082/1-390761711).References
- J.-F. Lutz, J.-M. Lehn, E. W. Meijer and K. Matyjaszewski, Nat. Rev. Mater., 2016, 1, 16024 CrossRef CAS
.
- A. S. Abd-El-Aziz, M. Antonietti, C. Barner-Kowollik, W. H. Binder, A. Böker, C. Boyer, M. R. Buchmeiser, S. Z. D. Cheng, F. D'Agosto, G. Floudas, H. Frey, G. Galli, J. Genzer, L. Hartmann, R. Hoogenboom, T. Ishizone, D. L. Kaplan, M. Leclerc, A. Lendlein, B. Liu, T. E. Long, S. Ludwigs, J. F. Lutz, K. Matyjaszewski, M. A. R. Meier, K. Müllen, M. Müllner, B. Rieger, T. P. Russell, D. A. Savin, A. D. Schlüter, U. S. Schubert, S. Seiffert, K. Severing, J. B. P. Soares, M. Staffilani, B. S. Sumerlin, Y. Sun, B. Z. Tang, C. Tang, P. Théato, N. Tirelli, O. K. C. Tsui, M. M. Unterlass, P. Vana, B. Voit, S. Vyazovkin, C. Weder, U. Wiesner, W. Y. Wong, C. Wu, Y. Yagci, J. Yuan and G. Zhang, Macromol. Chem. Phys., 2020, 221, 1–22 CrossRef
.
- M. Karayianni and S. Pispas, J. Polym. Sci., 2021, 59, 1874–1898 CrossRef CAS
.
- U. Tritschler, S. Pearce, J. Gwyther, G. R. Whittell and I. Manners, Macromolecules, 2017, 50, 3439–3463 CrossRef CAS
.
- G. Verma and P. A. Hassan, Phys. Chem. Chem. Phys., 2013, 15, 17016–17028 RSC
.
- Y. Mai, Z. An and S. Liu, Macromol. Rapid Commun., 2022, 43, 10–12 CrossRef
.
- S. C. Owen, D. P. Y. Chan and M. S. Shoichet, Nano Today, 2012, 7, 53–65 CrossRef CAS
.
- X. Jin, P. Sun, G. Tong and X. Zhu, Biomaterials, 2018, 178, 738–750 CrossRef CAS
.
- X. Fan, Z. Li and X. J. Loh, Polym. Chem., 2016, 7, 5898–5919 RSC
.
- J. M. Ren, T. G. McKenzie, Q. Fu, E. H. H. Wong, J. Xu, Z. An, S. Shanmugam, T. P. Davis, C. Boyer and G. G. Qiao, Chem. Rev., 2016, 116, 6743–6836 CrossRef CAS
.
- D. A. Resendiz-Lara, S. Azhdari, H. Gojzewski, A. H. Gröschel and F. R. Wurm, Chem. Sci., 2023, 14, 11273–11282 RSC
.
- T. Pelras, C. S. Mahon and M. Müllner, Angew. Chem., Int. Ed., 2018, 57, 6982–6994 CrossRef CAS
.
- G. Xie, M. R. Martinez, M. Olszewski, S. S. Sheiko and K. Matyjaszewski, Biomacromolecules, 2019, 20, 27–54 CrossRef CAS PubMed
.
- A. L. Liberman-Martin, C. K. Chu and R. H. Grubbs, Macromol. Rapid Commun., 2017, 38, 1–15 CrossRef
.
- I. Lapkriengkri, K. R. Albanese, A. Rhode, A. Cunniff, A. A. Pitenis, M. L. Chabinyc and C. M. Bates, Annu. Rev. Mater. Res., 2024, 54, 27–46 CrossRef CAS
.
- D. Saha, C. L. Witt, R. Fatima, T. Uchiyama, V. Pande, D. P. Song, H. F. Fei, B. M. Yavitt and J. J. Watkins, ACS Nano, 2025, 19, 1884–1910 CrossRef CAS PubMed
.
- G. Xie, M. R. Martinez, M. Olszewski, S. S. Sheiko and K. Matyjaszewski, Biomacromolecules, 2019, 20, 27–54 CrossRef CAS
.
- T. Pan, S. Dutta, Y. Kamble, B. B. Patel, M. A. Wade, S. A. Rogers, Y. Diao, D. Guironnet and C. E. Sing, Chem. Mater., 2022, 34, 1990–2024 CrossRef CAS
.
- K. Hu, R. Mu, Z. Ma and B. Li, Macromol. Rapid Commun., 2025, 46, 1–35 Search PubMed
.
- M. Müllner, Chem. Commun., 2022, 5683–5716 RSC
.
- G. Alberti, C. Zanoni, V. Losi, L. R. Magnaghi and R. Biesuz, Chemosensors, 2021, 9, 108 CrossRef CAS
.
- A. Reisch and A. S. Klymchenko, Small, 2016, 12, 1968–1992 CrossRef CAS
.
- C. Reichardt, Chem. Rev., 1994, 94, 2319–2358 CrossRef CAS
.
- J. Zhang, Y. Yao, Y. Zhang, D. Wu, W. Li, A. K. Whittaker and A. Zhang, Macromolecules, 2023, 56, 3931–3944 CrossRef CAS
.
- X. Qin, X. Yang, L. Du and M. Li, RSC Med. Chem., 2021, 12, 1826–1838 RSC
.
- Z. Zhang, Z. Li, Y. Shi and Y. Chen, Langmuir, 2024, 40, 7286–7299 CrossRef CAS
.
- M. Müllner, Macromol. Chem. Phys., 2016, 217, 2209–2222 CrossRef
.
- Z. Zheng, M. Müllner, J. Ling and A. H. E. Müller, ACS Nano, 2013, 7, 2284–2291 CrossRef CAS PubMed
.
- B. S. Sumerlin, D. Neugebauer and K. Matyjaszewski, Macromolecules, 2005, 38, 702–708 CrossRef CAS
.
- D. Neugebauer, B. S. Sumerlin, K. Matyjaszewski, B. Goodhart and S. S. Sheiko, Polymer, 2004, 45, 8173–8179 CrossRef CAS
.
- F. C. Giacomelli, P. Stepánek, C. Giacomelli, V. Schmidt, E. Jäger, A. Jäger and K. Ulbrich, Soft Matter, 2011, 7, 9316–9325 RSC
.
- K. Liang, S. T. Gunawan, J. J. Richardson, G. K. Such, J. Cui and F. Caruso, Adv. Healthcare Mater., 2014, 3, 1551–1554 CrossRef CAS
.
- M. Müllner, J. Yuan, S. Weiss, A. Walther, M. Förtsch, M. Drechsler and A. H. E. Müller, J. Am. Chem. Soc., 2010, 132, 16587–16592 CrossRef
.
- S. Dhar, S. Singha Roy, D. K. Rana, S. Bhattacharya, S. Bhattacharya and S. C. Bhattacharya, J. Phys. Chem. A, 2011, 115, 2216–2224 CrossRef CAS PubMed
.
- R. M. Duke, E. B. Veale, F. M. Pfeffer, P. E. Kruger and T. Gunnlaugsson, Chem. Soc. Rev., 2010, 39, 3936–3953 RSC
.
- G. Loving and B. Imperiali, J. Am. Chem. Soc., 2008, 130, 13630–13638 CrossRef CAS PubMed
.
- S. S. Bag, M. K. Pradhan, R. Kundu and S. Jana, Bioorg. Med. Chem. Lett., 2013, 23, 96–101 CrossRef CAS
.
- S. M. Hickey, I. R. D. Johnson, E. Dallerba, M. J. Hackett, M. Massi, J. Lazniewska, L. A. Thurgood, F. M. Pfeffer, D. A. Brooks and T. D. Ashton, Dyes Pigm., 2023, 217, 111382 CrossRef CAS
.
- F. Meng, J. He, J. Niu, Y. Li, P. Gao and X. Yu, J. Mater. Chem. B, 2022, 10, 8875–8882 RSC
.
- B. Dong, X. Song, X. Kong, C. Wang, N. Zhang and W. Lin, J. Mater. Chem. B, 2017, 5, 988–995 RSC
.
- L. Salminen, E. Karjalainen, V. Aseyev and H. Tenhu, Langmuir, 2022, 38, 5135–5148 CrossRef CAS
.
- Y. Q. Hu, M. S. Kim, B. S. Kim and D. S. Lee, Polymer, 2007, 48, 3437–3443 CrossRef CAS
.
- V. Bütün, S. P. Armes and N. C. Billingham, Polymer, 2001, 42, 5993–6008 CrossRef
.
- M. E. Graziotto, L. D. Adair, A. Kaur, P. Vérité, S. R. Ball, M. Sunde, D. Jacquemin and E. J. New, RSC Chem. Biol., 2021, 2, 1491–1498 RSC
.
- K. G. Leslie, D. Jacquemin, E. J. New and K. A. Jolliffe, Chem.–Eur. J., 2018, 24, 5569–5573 CrossRef CAS
.
-
L. D. Adair, N. K. Tan and E. J. New, Molecular Fluorescent Sensors for Cellular Studies, 2022, pp. 129–172 Search PubMed
.
- N. Kongkatigumjorn, S. A. Smith, M. Chen, K. Fang, S. Yang, E. R. Gillies, A. P. R. Johnston and G. K. Such, ACS Appl. Nano Mater., 2018, 1, 3164–3173 CrossRef CAS
.
- S. O. Catt, C. Vazquez-Martel and E. Blasco, Mol. Syst. Des. Eng., 2025, 10, 176–183 RSC
.
- B. Weidinger, G. Yang, N. von Coelln, H. Nirschl, I. Wacker, P. Tegeder, R. R. Schröder and E. Blasco, Adv. Sci., 2023, 10, 2302756 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |