Open Access Article
Open Access ArticleNavigating the next frontier in biomedicine: breakthroughs and insights in nucleic acid therapeutics
Shanchao
Wu
a,
Zhihui
Zhang
a,
Zilong
Zhao
 a,
Cheng
Cui
*a and
Weihong
Tan
a,
Cheng
Cui
*a and
Weihong
Tan
 *ab
*ab
aMolecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, College of Biology, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China. E-mail: ccui@hnu.edu.cn
bZhejiang Cancer Hospital, Hangzhou Institute of Medicine (HIM), Chinese Academy of Sciences, Hangzhou, Zhejiang 310022, China. E-mail: tan@him.cas.cn
First published on 13th January 2026
Abstract
Nucleic acid therapeutics are rapidly emerging as a transformative drug paradigm, offering precise and programmable regulation of gene expression across a broad spectrum of diseases. This review summarizes recent advances in key platforms—including antisense oligonucleotides, siRNA, miRNA, mRNA, and aptamers—emphasizing their unique mechanisms of action and therapeutic potential. We systematically outline critical contributions of chemical modification and delivery engineering, including backbone and sugar modifications, site-specific design, N-acetylgalactosamine (GalNAc) conjugation, and lipid nanoparticles, which collectively enhance stability, target specificity, and clinical applicability. Finally, we discuss persistent challenges such as immune activation, large-scale manufacturing, and long-term safety, and provide perspectives on future directions involving CRISPR-based gene editing, synthetic biology, nanotechnology, smart delivery systems, and combination therapies, aiming to offer strategic insights for the development and clinical translation of nucleic acid drugs.
1. Introduction
The central dogma of molecular biology, a cornerstone of modern biology, highlights the critical role of nucleic acids in carrying genetic information.1 These fundamental biomolecules are ubiquitous across all living organisms and govern essential life processes, such as growth, heredity, and variation.2,3 With the advancement in molecular biology, nucleic acid-based therapeutics has emerged as a promising strategy for targeting pathogenic genes or mRNA, opening new avenues for disease treatment.4Nucleic acid therapeutics leverage the sequence specificity and regulatory capacity of nucleic acids to influence gene expression and translation, enabling precise intervention through recognition of endogenous nucleic acid sequences.5 Since the 1950s, breakthroughs in this field have been repeatedly honored with Nobel Prizes, underscoring both their scientific significance and clinical value. The elucidation of the DNA double-helix structure by Watson and Crick in 1953 established the molecular foundation for rational drug design.6 Subsequently, the identification of catalytic RNAs (ribozymes) revealed that nucleic acids could serve not only as carriers of genetic information but also as functional biomolecules, inspiring the development of riboswitches and, ultimately, nucleic acid aptamers as versatile molecular tools for gene regulation and therapeutic applications. Building on these advances, groundbreaking research in genetics led to the 2002 Nobel Prize, which deepened our understanding of genetic regulation in processes such as organogenesis and programmed cell death.7 Parallel to these biological insights, the emergence of nucleic acid nanostructures introduced a new structural dimension to the field. In 1996, Chad A. Mirkin pioneered spherical nucleic acids (SNAs)—nucleic acid shells densely arranged on nanoparticle cores—establishing a new paradigm for programmable nanomaterials.8 A decade later, in 2006, Paul W. K. Rothemund introduced DNA origami, demonstrating the precise folding of long DNA strands into two- and three-dimensional architectures.9 These breakthroughs laid the foundation for the development of multifunctional DNA/RNA nanostructures with programmable shapes, spatial addressability, and biomedical applications, marking a transformative expansion of nucleic acid science beyond sequence information. In the 21st century, the discovery of RNA interference (2006) catalyzed the development of siRNA-based drugs, with the approval of Onpattro (patisiran) in 2018 as the first RNAi therapy, demonstrating the clinical feasibility of nucleic acid medicines.10 The 2020 Nobel Prize awarded to CRISPR-Cas9, a gene-editing tool guided by RNA, further accelerated nucleic acid delivery and genome editing in vivo.11 In 2023, the Nobel Prize recognized the role of nucleoside modifications in enhancing mRNA translation while evading immune recognition, laying the foundation for the success of mRNA vaccines.12 In 2024, the Nobel Prize further acknowledged the therapeutic potential of microRNAs in post-transcriptional gene regulation, broadening the therapeutic landscape of nucleic acid-based therapies.13 Collectively, these accolades not only affirm the extraordinary progress of nucleic acid science but also emphasize the transformative potential of nucleic acid therapeutics in the era of precision medicine (Fig. 1).14,15
Since the 1980s, the target-based approach to drug discovery has matured, driving the development of numerous innovative therapeutics.16–18 Conventional small-molecule drugs and antibody-based biologics typically exert therapeutic effects by binding to target proteins such as enzymes, receptors, or ion channels.19 Small molecules offer advantages including facile synthesis, oral bioavailability, favorable pharmacokinetic properties, and efficient membrane permeability.20 However, their development is severely constrained by target “druggability.” Among the ≈20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 protein-coding genes in the human genome, only about 3000 are considered druggable, with just ≈700 yielding approved drugs.21 Antibody therapeutics, by contrast, can target a wider array of proteins and can be engineered to improve their affinity and safety.22,23 Nonetheless, their clinical application is limited by structural complexity, high manufacturing costs, and the need for parenteral administration.24 Moreover, antibodies generally act only on extracellular or cell-surface proteins, significantly restricting their therapeutic scope.25–27 In contrast, nucleic acid therapeutics offer unique advantages. They regulate gene expression through base-pair complementarity rather than direct protein binding, bypassing the limitations of protein “druggability”. Furthermore, with appropriate delivery systems, nucleic acids can penetrate cells and act intracellularly, enabling broad regulation of intracellular, extracellular, and membrane-associated targets. Therapeutic nucleic acids can be designed rapidly based on known target gene sequences, with chemical modifications and delivery strategies developed independently. These capabilities position nucleic acid therapeutics as a transformative approach in precision medicine, offering novel solutions for both common and rear diseases, and overcoming the inherent limitations of traditional drug discovery.
000 protein-coding genes in the human genome, only about 3000 are considered druggable, with just ≈700 yielding approved drugs.21 Antibody therapeutics, by contrast, can target a wider array of proteins and can be engineered to improve their affinity and safety.22,23 Nonetheless, their clinical application is limited by structural complexity, high manufacturing costs, and the need for parenteral administration.24 Moreover, antibodies generally act only on extracellular or cell-surface proteins, significantly restricting their therapeutic scope.25–27 In contrast, nucleic acid therapeutics offer unique advantages. They regulate gene expression through base-pair complementarity rather than direct protein binding, bypassing the limitations of protein “druggability”. Furthermore, with appropriate delivery systems, nucleic acids can penetrate cells and act intracellularly, enabling broad regulation of intracellular, extracellular, and membrane-associated targets. Therapeutic nucleic acids can be designed rapidly based on known target gene sequences, with chemical modifications and delivery strategies developed independently. These capabilities position nucleic acid therapeutics as a transformative approach in precision medicine, offering novel solutions for both common and rear diseases, and overcoming the inherent limitations of traditional drug discovery.
2. Major classes of nucleic acid therapeutics and their recent clinical advances
2.1. Antisense oligonucleotides (ASOs)
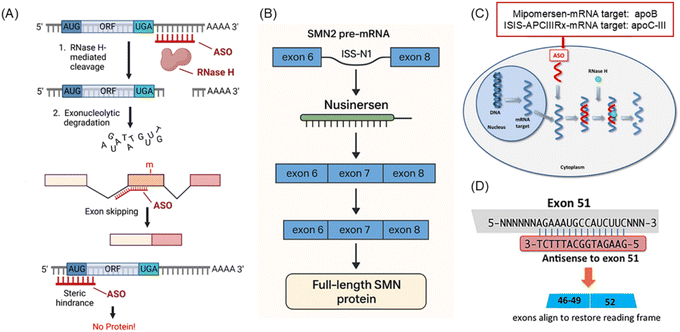
Splice reprogramming refers to the modulation of pre-mRNA splicing within the nucleus, thereby altering the composition of mature mRNA. ASOs can bind to critical regions of pre-mRNA and, by blocking the interaction of inhibitory splicing factors or recruiting activators, promote the inclusion or skipping of specific exons to achieve selective splicing.35 In 2013, Singh et al. first reported the therapeutic application of this mechanism: in cells derived from patients with spinal muscular atrophy (SMA), ASOs targeting the 3′-end of the intronic structure ISTL1 effectively corrected the exon-splicing defect of the SMN2 gene.36 In addition, ASOs can also exert therapeutic effects by targeting long non-coding RNAs (lncRNAs). For example, in β-thalassemia, ASOs directed against an antisense lncRNA of the BCL11A gene were shown to increase fetal hemoglobin (HbF) expression by approximately 40%.37 Furthermore, Yang et al. demonstrated that the lncRNA HIF1A-AS2 is regulated by the oncogene KRAS in lung cancer and promotes the proliferation of NSCLC. Inhibition of HIF1A-AS2 using ASOs markedly enhanced tumor sensitivity to both the MYC inhibitor 10058-F4 and cisplatin treatment.38
| Drug name | Brand names | Approval year | Indication |
|---|---|---|---|
| Fomivirsen | Vitravene | 1998 | CMV retinitis in AIDS patients |
| Mipomersen | Kynamro | 2013 | Homozygous familial hypercholesterolemia (HoFH) |
| Nusinersen | Spinraza | 2016 | Spinal muscular atrophy (SMA) |
| Eteplirsen | Exondys | 2016 | Duchenne muscular dystrophy (DMD), exon 51 skipping |
| Inotersen | Tegsedi | 2018 | Hereditary transthryetin-mediated amyloidosis (hATTR-PN) |
| Volanesorsen | Waylivra | 2019 | Familial chylomicronemia syndrome (FCS) |
| Golodirsen | Vyondys 53 | 2019 | DMD, exon 53 skipping |
| Viltolarsen | Viltepso | 2020 | DMD, exon 53 skipping |
| Casimersen | Amondy 45 | 2021 | DMD, exon 45 skipping |
| Tofersen | Qalsody | 2023 | SOD1 mutration-associated amytrophic lateral sclerosis (ALS) |
Nusinersen (Spinraza) is an ASO approved for the treatment of SMA. SMA is primarily caused by mutations in the survival motor neuron 1 (SMN1), leading to deficiency of the SMN protein.40 Nusinersen is a splice-modulating ASO that specifically binds to the intronic splicing silencer N1 (ISS-N1) region of SMN2 pre-mRNA. SMN2 is a gene highly homologous to SMN1, but its transcripts typically undergo exon 7 skipping, resulting in truncated and nonfunctional proteins.41 By blocking the binding of inhibitory splicing factors to ISS-N1, and potentially recruiting activators, nusinersen promotes the inclusion of exon 7 in SMN2 pre-mRNA, thereby significantly increasing the production of full-length, functional SMN protein (Fig. 2B).42 The drug is delivered via intrathecal injection, enabling it to bypass the blood–brain barrier and reach the cerebrospinal fluid to target spinal motor neurons. Multiple pivotal clinical trials have demonstrated that nusinersen markedly improves motor function, survival, and respiratory capacity in SMA patients, with particularly pronounced efficacy in presymptomatic and early-onset infants.43 As the first approved therapy for SMA, nusinersen has fundamentally altered the natural course of the disease.44
Mipomersen (Kynamro) is an antisense oligonucleotide that targets the Apolipoprotein B-100 (ApoB-100) mRNA and is approved for the treatment of homozygous familial hypercholesterolemia (HoFH). HoFH results from mutations in the ApoB-100 gene, leading to loss of low-density lipoprotein (LDL) receptor function. ApoB-100 is a structural protein required for the hepatic synthesis of very-low-density lipoprotein (VLDL), which is subsequently metabolized into LDL.45 Mipomersen is a 2′-O-methoxyethyl (2′-MOE)-modified Gapmer ASO that promotes the degradation of ApoB-100 mRNA via RNase H1 activation.46 Following subcutaneous administration, the drug inhibits hepatic ApoB-100 synthesis and reduces VLDL secretion, thereby significantly lowering plasma LDL cholesterol levels (Fig. 2C).47 However, its use is associated with hepatotoxicity risks inhibition of VLDL secretion leads to triglyceride accumulation in hepatocytes, resulting in elevated transaminase levels ≥3× the upper limit of normal (ULN) in approximately 10–15% of patients.
Eteplirsen is an antisense oligonucleotide developed for the treatment of Duchenne muscular dystrophy (DMD). DMD is caused by mutations in the DMD gene, resulting in loss of dystrophin protein, which disrupts muscle membrane stability and leads to progressive muscle degeneration.48 Patients typically lose ambulation before the age of 12 and die from cardiorespiratory failure before age 20. Eteplirsen is an exon-skipping ASO designed for patients with mutations amenable to exon 51 skipping. These mutations induce a frameshift and premature stop codons, yielding nonfunctional ytruncated proteins. Eteplirsen belongs to the class of phosphorodiamidate morpholino oligomers (PMOs) and targets the splicing enhancer within exon 51 of DMD pre-mRNA. By binding to this regulatory region, it creates steric hindrance and blocks the recognition of exon 51 by the splicing factor U1 snRNP, thereby inducing exon 51 skipping. This allows exons 50 and 52 to be joined directly, restoring the reading frame and producing a shortened but partially functional Becker-type dystrophin protein (Fig. 2D).49 Clinical trials demonstrated increased dystrophin expression in muscle biopsies from eteplirsen-treated patients, along with delayed decline in the six-minute walk test.50 Notably, eteplirsen became the first drug in history to be conditionally approved for DMD based on a surrogate endpoint.
2.1.3.1. Backbone modifications. Phosphorothioate (PS) modification involves replacing the non-bridging oxygen atom of the phosphate backbone with sulfur. PS linkages exist as two stereoisomers, whereas natural phosphodiester bonds are prochiral. PS-ASOs synthesized via conventional methods generally consist of mixtures of diastereomers, with certain stereoisomers displaying higher activity. This modification has become one of the most widely applied chemical strategies.52 Phosphoramidate (PN) modification substitutes oxygen with nitrogen to form a P–N bond, while phosphorodiamidate borane (PB) modification introduces a tetrahedral structure via boron incorporation, which can increase the melting temperature (Tm) by approximately 8 °C; these modifications are currently in preclinical development. Such backbone modifications significantly enhance nuclease resistance and prolong the in vivo half-life of ASOs. Moreover, the negative charge facilitates binding to plasma proteins (e.g., albumin), thereby improving pharmacokinetics, enhancing tissue distribution, and promoting cellular uptake.53 Nevertheless, these modifications may reduce RNA-binding affinity and increase nonspecific protein interactions, potentially leading to adverse effects such as thrombocytopenia or nephrotoxicity. Phosphorodiamidate morpholino oligomers (PMOs) represent another backbone modification strategy, in which the ribose sugar is replaced by a morpholine ring and phosphorodiamidate linkages are introduced. This modification not only enhances nuclease resistance but also confers minimal immune activation potential.
2.1.3.2. Sugar modifications. Major sugar ring modifications include 2′-OMe, 2′-MOE, locked nucleic acid (LNA), and 2′-fluoro (2′-F). The 2′-OMe modification improves nuclease resistance, increases binding affinity (as reflected by elevated Tm values), and reduces immunogenicity. The 2′-MOE modification further increases hydrophobicity, resulting in superior nuclease resistance, higher binding affinity, and lower immunogenicity; however, the increased molecular weight may impair in vivo delivery efficiency. LNA modifications introduce a rigid methylene bridge between the 2′-O and 4′-C positions, markedly enhancing both binding affinity and nuclease stability.54,55 LNA has been widely applied to enhance the potency of short ASOs; however, potential hepatotoxicity—particularly associated with TGC/TCC motifs—remains a safety concern. Yoshida et al.56 systematically screened 17 nucleobase derivatives and 4 novel modifications, identifying several that significantly reduced the hepatotoxicity of LNA-ASOs (Fig. 3B). The 2′-F modification also provides improvements in nuclease stability and binding affinity,57 although it may partially inhibit the activation efficiency of RNase H.
The terminal nucleotides of ASOs can be functionalized via conjugation strategies with specific ligands (such as GalNAc, cholesterol, peptides, or antibody fragments), thereby enabling active targeting of cell surface receptors. This approach markedly enhances cellular uptake efficiency, reduces systemic dosing requirements, and minimizes off-target effects and toxicity. Among these, GalNAc-ASO conjugates efficiently target the asialoglycoprotein receptor (ASGPR) and have been extensively applied for liver-specific delivery.58,59 In terms of structural design, Gapmers integrate multiple modification advantages: the central region, typically composed of 8–10 DNA or phosphorothioate-DNA nucleotides, is responsible for recruiting RNase H1, whereas the flanking wings are modified with 2′-OMe, 2′-MOE, or LNA nucleotides to enhance binding affinity and stability. Seth and colleagues reported a site-specific incorporation strategy in which 5′-methyl DNA nucleotide stereoisomers were introduced into the Gapmer region.60 Their systematic evaluation demonstrated that placing such modifications at the third and fourth positions enhanced the therapeutic performance of PS-ASOs while modulating cytotoxicity, highlighting the clinical potential of this design. It should be noted, however, that ASO modifications must be rationally balanced: excessive modifications (e.g., full phosphorothioation or an abundance of high-affinity substitutions) may lead to overly strong binding, off-target effects, or increased toxicity.
2.2. MicroRNA
In addition, Yuan and colleagues78 developed a multivariate-gated catalytic hairpin assembly (CHA) nanosensor that enabled specific amplified imaging of miR-21 in colorectal cancer tissues (Fig. 4D) and further demonstrated that miR-21 contributes to colorectal tumorigenesis by suppressing the expression of the mismatch repair protein hMSH2. Conversely, tumor-suppressive miRNAs (TSmiRs) are frequently downregulated or lost in tumors, where their physiological role is to inhibit the expression of oncogenes. Thus, restoration of TSmiR expression is considered to have therapeutic potential. The let-7 family represents one of the earliest discovered TSmiRs, and reduced expression levels have been strongly associated with shorter postoperative survival in cancer patients. Forced expression of let-7 in both in vitro and in vivo models effectively suppressed tumor growth (Fig. 4E). Let-7 exerts its tumor-suppressive effects primarily by directly targeting multiple oncogenes, including RAS, MYC, and HMGA2.79 Consequently, diminished expression of let-7 has been recognized as a prognostic biomarker for predicting survival outcomes in lung cancer patients.80
Conversely, miRNA mimics are designed to supplement or restore the function of tumor-suppressive miRNAs. To enhance stability and delivery efficiency, they are typically modified with 2′-OMe or PS chemistries. These double-stranded molecules are often encapsulated in lipid nanoparticles (LNPs) or other delivery vehicles, which protect their duplex structure, facilitate cellular uptake, and promote endosomal escape. A representative example is MRX34, a liposomal miR-34a mimic, which was the first miRNA-based therapeutic developed for cancer. miR-34a, the major member of the miR-34 family, is transcriptionally regulated by the tumor suppressor p53, which is frequently mutated or deleted in cancers. In most malignancies, miR-34a is downregulated (Fig. 4F).84 MRX34 exhibited antitumor activity in phase I clinical trials across various solid tumors, reducing the expression of miR-34 target genes, oncogenes, and immune evasion-related genes. However, its development was ultimately discontinued due to severe immune-related adverse events.85,86
2.3. siRNA
Following administration, LNP–siRNA is primarily internalized into hepatocytes through ApoE coating and subsequent interaction with LDLR, whereas GalNAc–siRNA undergoes ASGPR-mediated endocytosis. After particles or conjugates enter early endosomes, endosomal escape represents a critical bottleneck for pharmacological activity: LNPs rely on ionizable lipids that disrupt membranes under acidic conditions to facilitate escape, while GalNAc–siRNAs depend on limited spontaneous leakage into the cytoplasm.93,94 Once in the cytoplasm, double-stranded siRNA is incorporated into RISC: Ago2 recognizes and retains the thermodynamically less stable strand as the guide strand (whose 5′ phosphate pairs with the MID domain), while the passenger strand is either cleaved or displaced. The guide strand then uses its seed region (nt 2–8) to search for complementary sequences and establishes full-length base pairing with the target mRNA.95 Ago2 subsequently cleaves the mRNA backbone between the 10th and 11th nucleotides relative to the guide strand's 5′ end, with the cleavage products degraded by cellular ribonucleases, while RISC undergoes multiple catalytic cycles to mediate gene silencing (Fig. 5C). Meanwhile, partial complementarity may elicit miRNA-like off-target effects, most commonly via seed pairing at the 3′ UTR.96 To mitigate this, modern siRNA design employs site-selective base and sugar modifications, together with thermodynamic asymmetry optimization, to suppress off-target activity and extend half-life. Benefiting from the efficient turnover of the Ago2–RISC complex with target mRNAs in hepatocytes, coupled with enhanced chemical stability, clinical applications have now achieved durable gene silencing with quarterly to semiannual dosing regimens, offering a practical therapeutic strategy for long-term management of chronic diseases.
The success of siRNA therapeutics is fundamentally supported by an integrated engineering framework encompassing “chemical modification, receptor–ligand targeting, and endosomal escape”.103,104 At the sequence level, the prevailing clinical strategy adopts alternating patterns of 2′-OMe and 2′-F ribose modifications, combined with limited terminal PS linkages.105 This configuration enhances nuclease resistance and plasma stability while markedly attenuating innate immune recognition. Notably, the incorporation of 2′-OMe at U-rich motifs effectively suppresses TLR7/8-mediated cytokine release, representing a classical approach to reducing immunostimulatory reactivity (Fig. 5D).106 To further improve Ago2 loading and prolong in vivo exposure, the guide strand 5′ terminus is frequently modified with a 5′-(E)-vinylphosphonate group, which mimics the natural 5′-phosphate, strengthens binding to the MID domain, and thereby enhances pharmacological activity and tissue retention. In addition, site-specific modifications within the seed region (nt 2–8), combined with thermodynamic asymmetry design, mitigate miRNA-like off-target effects and hepatotoxicity, as exemplified by Alnylam's Enhanced Stabilization Chemistry (ESC) and Enhanced Stabilization Chemistry Plus (ESC+) platforms. At the delivery level, early-generation LNP systems (e.g., the MC3 ionizable lipid used in patisiran) rely on lipid protonation and phase transition upon endosomal acidification to facilitate endosomal escape.107–109 In contrast, triantennary GalNAc conjugates exploit ASGPR-mediated endocytosis in hepatocytes, thereby enabling subcutaneous administration, liver-specific uptake, and quarterly to biannual dosing intervals. This strategy has been successfully validated in multiple approved products, including givosiran, lumasiran, vutrisiran, and inclisiran.
2.4. Aptamer
Aptamers exhibit dual mechanisms in therapeutic applications, the first being their role as targeted delivery vehicles. Through covalent or noncovalent conjugation with therapeutic agents—including small-molecule drugs, toxins,116 radionuclides, siRNA/ASO,117 and proteins118—aptamers enable precise delivery. In 2009, Huang et al. first introduced the concept of aptamer–drug conjugates (ApDCs) (Fig. 6A), which has since become the most widely adopted strategy for utilizing aptamers as delivery tools.119 For instance, Bagalkot et al.120 demonstrated the physical conjugation of an RNA aptamer with doxorubicin (DOX) via intercalation of the anthracyclic ring, enabling targeted delivery to prostate-specific membrane antigen (PSMA)-positive cancer cells, while Li et al.121 designed a cathepsin B-responsive dipeptide linker (NucA-PTX) that releases paclitaxel intracellularly upon enzymatic cleavage, thereby achieving tumor-selective drug delivery (Fig. 6B). The hydrophilic backbone of aptamers enhances the solubility of such conjugates, promotes tumor accumulation, improves therapeutic efficacy, and reduces systemic toxicity. During the delivery process, aptamers specifically bind to receptors on the surface of target cells and undergo receptor-mediated endocytosis, after which active drugs are released within endosomal/lysosomal microenvironments (e.g., low pH, reductive conditions, or enzymatic cleavage) or via endosomal escape mechanisms,122 thereby exerting intracellular therapeutic effects while markedly reducing off-target toxicity.123 Moreover, aptamers can also serve as functional carriers for photosensitizers. For example, Tan and colleagues124 developed giant membrane vesicles (GMVs) co-loaded with the aptamer AS1411, the photosensitizer TMPyP4, and the photothermal agent ICG, using chol-Sgc8 aptamer for PTK7 targeting (Fig. 6C), which demonstrated enhanced cytotoxicity and therapeutic efficacy against CCRF-CEM cells.
The second therapeutic mechanism of aptamers lies in their direct function as modulators (antagonists or agonists), whereby they regulate the biological activity of target proteins through binding to specific functional domains. Aptamers can exploit complex secondary structures such as G-quadruplexes to precisely recognize protein epitopes: these structures are formed by guanine-rich sequences that generate planar G-quartets via Hoogsteen hydrogen bonding, which further stack into stable G-quadruplexes stabilized by monovalent cations within the central channel (Fig. 6D).125 The G-quadruplex provides a rigid, negatively charged interface that enables high-affinity and high-specificity recognition of positively charged surface regions or functional pockets of proteins, thereby directly interfering with protein function. As antagonists, aptamers block the interaction of target proteins with their natural ligands or receptors to inhibit downstream signaling. For example, the G-quadruplex-forming aptamer AS1411 binds to nucleolin, which is overexpressed on cancer cell surfaces, thereby suppressing its function and inducing apoptosis and cell cycle arrest.126 Conversely, as agonists, aptamers activate signaling by binding to and stabilizing active conformations of their targets. Although currently limited in number, reported examples include RNA aptamers against HER3,127 CD28,128 OX40,129 4-1BB,130 CD40,131 VEGFR-2,132 and the insulin receptor (IR).133
2.5. mRNA
Messenger RNA (mRNA) represents a versatile platform for nucleic acid therapeutics. Beyond the remarkable success of mRNA vaccines, its applications have expanded to encompass diverse therapeutic modalities, including protein replacement therapy, gene therapy, and regenerative medicine. Therefore, this section provides an overview of the major therapeutic strategies based on mRNA technology, highlighting their underlying mechanisms and recent clinical progress.2.5.1.1. Mechanism of action of mRNA vaccine. The concept of using mRNA as a vaccine substrate dates back to 1990, when Wolff et al. demonstrated for the first time that intramuscular injection of mRNA/DNA into mice could successfully express reporter proteins, thereby proving the feasibility of “in vivo translation of messenger RNA” and laying the foundation for mRNA therapeutics.144,145 However, the intrinsic immunogenicity and instability of mRNA long hindered its clinical development. Around 2005, Karikó and Weissman introduced nucleoside modifications (e.g., Ψ and 1-methylpseudouridine) that markedly reduced innate immune activation and enhanced translational efficiency, a breakthrough widely recognized as the turning point for the platform.146 This work, which paved the way for COVID-19 mRNA vaccines, was honored with the 2023 Nobel Prize in Physiology or Medicine. In 2020, mRNA vaccines BNT162b2 and mRNA-1273 were granted emergency use authorization and subsequently full approval worldwide, marking the successful transition of mRNA vaccines “from concept to industry.”147,148
The fundamental composition of mRNA vaccines consists of two major components: the nucleic acid sequence itself and the delivery system. The mRNA molecule typically contains a 5′-cap structure, untranslated regions (UTRs), an open reading frame (ORF), and a 3′ polyadenylated tail (poly-A tail), which collectively ensure intracellular stability and efficient translation. To overcome nuclease degradation and the cell membrane barrier, mRNA is encapsulated within lipid nanoparticles (LNPs) for delivery. A typical LNP is composed of four essential constituents: (i) ionizable lipids, which electrostatically complex with negatively charged mRNA under acidic conditions and facilitate endosomal release; (ii) cholesterol, which enhances particle stability and membrane fluidity; (iii) structural phospholipids (e.g., DSPC), which maintain bilayer integrity; and (iv) PEG–lipids, which form a hydrophilic corona on the particle surface to reduce plasma protein adsorption and prolong circulation (Fig. 7A).149
Following intramuscular injection, LNP-encapsulated mRNA forms a transient local depot, with a portion of particles draining to regional lymph nodes (Fig. 7B).150,151 The combination of mRNA with ionizable lipids triggers mild innate immune signalling, recruiting antigen-presenting cells (APCs) such as dendritic cells (DCs). Upon uptake of LNPs by myocytes and APCs, endosomal acidification leads to protonation of ionizable lipids, disrupting membrane integrity and promoting cytosolic release of mRNA, which is subsequently translated by ribosomes. The resulting antigens undergo processing through the endoplasmic reticulum-Golgi network, appearing either as secreted proteins or membrane-anchored forms. Secreted antigens are taken up by APCs and presented via the MHC II pathway to activate CD4+ T cells (particularly T follicular helper cells, Tfh), driving germinal center reactions, class switching, and affinity maturation. In parallel, membrane-associated or endogenously synthesized antigens are processed by the immunoproteasome–TAP complex and presented via MHC I, thereby activating CD8+ T cells to mount cytotoxic responses.152
These processes generate neutralizing antibodies from 9 long-lived plasma cells in the bone marrow, together with memory B and T cells that establish durable protection. Antigen expression typically peaks within 24–48 h and declines as mRNA is degraded; booster doses rapidly expand the memory pool and significantly increase antibody titers. Ultimately, mRNA is degraded by nucleases, and LNPs are cleared through the hepatobiliary pathway. Importantly, mRNA vaccines do not enter the nucleus and pose no risk of genomic integration, while common adverse events are generally self-limited, consisting of local erythema/swelling and transient systemic symptoms.153,154
2.5.1.2. Clinical progress of mRNA vaccine. In recent years (2024–2025), the mRNA platform has transitioned from a single emergency response against COVID-19 toward a “multi-disease, routine” development paradigm, achieving multiple breakthroughs in clinical settings. In the respiratory field, Moderna's RSV vaccine mRESVIA (mRNA-1345) was approved by the FDA in 2024 for adults aged ≥60 years,155 and in 2025 its indication was further expanded to high-risk adults aged 18–59, reflecting the establishment of a relatively mature regulatory pathway for this indication.156 In the same year, its seasonal influenza candidate mRNA-1010 demonstrated a 26.6% superiority over a licensed standard-dose influenza vaccine in a phase III study involving 40
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 participants,157 and based on these results the company initiated regulatory submission discussions, marking the first clear clinical evidence of superiority for mRNA vaccines in influenza.158 Beyond respiratory infectious diseases, Moderna's cytomegalovirus (CMV) vaccine mRNA-1647 is advancing in the phase III CMVictory (P301) trial, with primary endpoints focusing on seroconversion prevention, safety, and immunogenicity, representing one of the first “routine” mRNA vaccines with registration potential following COVID-19. In addition, an RABV-G mRNA vaccine for rabies virus glycoprotein has entered phase I clinical evaluation.
800 participants,157 and based on these results the company initiated regulatory submission discussions, marking the first clear clinical evidence of superiority for mRNA vaccines in influenza.158 Beyond respiratory infectious diseases, Moderna's cytomegalovirus (CMV) vaccine mRNA-1647 is advancing in the phase III CMVictory (P301) trial, with primary endpoints focusing on seroconversion prevention, safety, and immunogenicity, representing one of the first “routine” mRNA vaccines with registration potential following COVID-19. In addition, an RABV-G mRNA vaccine for rabies virus glycoprotein has entered phase I clinical evaluation.
In oncology, progress is being driven by personalized neoantigen vaccines. mRNA-4157 (V940) in combination with pembrolizumab has advanced from melanoma into multiple phase III trials in NSCLC and other indications,159 with early follow-up data demonstrating sustained benefit in recurrence and metastasis outcomes for high-risk melanoma, thereby providing clinical evidence of durable efficacy for the “vaccine plus checkpoint inhibitor” strategy (Fig. 7C).160 In parallel with these clinical efforts, formulation and delivery technologies are addressing critical limitations. Several studies have confirmed that lyophilized mRNA–LNP formulations can maintain physicochemical and immunological activity at 4–25 °C for extended periods, offering a promising means to reduce cold-chain dependence.161 Meanwhile, organ-selective LNPs designed for extrahepatic delivery—such as the selective organ targeting (SORT) strategy, which incorporates defined fractions of supplemental lipids to tune biodistribution, and its derivatives—have achieved programmable biodistribution to tissues such as the lung and kidney through advances in materials and formulation engineering, opening a “targeted delivery” window for next-generation prophylactic and therapeutic mRNA agents.162
Overall, the recent progress of mRNA is characterized by three parallel dimensions—indication expansion, accumulation of registrational evidence, and advances in delivery/stability engineering—which together consolidate the public health value of the vaccine platform while providing a pathway and toolkit for the clinical translation of therapeutic mRNA.
The use of mRNA to provide functional copies of missing or dysfunctional proteins offers a highly promising strategy for the treatment of monogenic metabolic disorders. By encapsulating mRNA encoding functional enzymes within lipid nanoparticles (LNPs) and delivering them to target cells, it is possible to restore the activity of key metabolic pathways in vivo, thereby correcting long-term metabolic impairments. For example, Ding et al. developed a pseudouridine (Ψ)-modified codon-optimized mRNA–LNP formulation encoding human methylmalonyl-CoA mutase (hMUT), the enzyme most frequently mutated in methylmalonic acidemia (MMA). This system achieved efficient protein expression and remarkable metabolic improvement in murine models, effectively reversing the pathological phenotype of MMA; the therapy has now advanced to clinical evaluation.166 Similarly, Koeberl D. et al. reported the interim results of a phase I/II clinical trial for propionic acidemia (PA), which systematically evaluated the safety and efficacy of mRNA-3927—a dual mRNA therapeutic candidate encoding PCCA and PCCB—thereby providing proof-of-concept evidence for precise treatment of biallelic enzymatic deficiencies.167 In addition, Yamazaki K. et al. developed an engineered hOTC-mRNA/LNP formulation (encoding human ornithine transcarbamylase, hOTC) that demonstrated a significant dose-dependent therapeutic response in an ornithine transcarbamylase deficiency (OTCD) mouse model, markedly improving survival rates.168 These findings collectively underscore the clinical potential of mRNA-based protein replacement therapy for rare inherited metabolic disorders.
In the field of regenerative medicine, mRNA technology promotes tissue repair and regeneration by transiently inducing the expression of regenerative and pro-healing factors. For instance, Zangi L. et al. first demonstrated that synthetic mRNA can drive efficient in vivo expression of vascular endothelial growth factor A (VEGF-A).171 When chemically modified VEGF-A mRNA was directly injected into the myocardium of a mouse model of myocardial infarction, it markedly induced the differentiation of cardiac progenitor cells into endothelial cells, stimulated the formation of functional neovasculature, and significantly improved cardiac performance. This pioneering study established both the conceptual and experimental foundation for the use of mRNA in regenerative medicine, highlighting its broad potential in promoting tissue repair and organ regeneration.
2.6. Nucleic acid nanostructures
The aforementioned classes of nucleic acid therapeutics (ASOs, siRNAs, miRNAs, mRNAs, and aptamers) primarily rely on sequence design and chemical modification strategies to achieve precise regulation of gene expression and therapeutic intervention. With the rapid advances in structural biology and nanotechnology, researchers have further sought to engineer nucleic acids in the spatial dimension, giving rise to a new class of artificial nucleic acid nanostructures characterized by high programmability and controllable self-assembly.172 These structures exploit the stringent base-pairing principles of nucleic acids (A–T/U and C–G) to achieve sequence-specific recognition among designed segments.173 Through precise intra- or intermolecular hybridization, they can assemble into complex higher-order architectures at the nanoscale, thereby endowing the system with well-defined geometries and tunable functionalities. Representative examples of such nucleic acid nanostructures include spherical nucleic acids and DNA origami, which have emerged as versatile platforms for structural innovation and functional modulation in nucleic acid therapeutics.174SNAs exhibit distinctive structural and functional advantages.177 First, their core–shell architecture endows the system with exceptional physicochemical stability.178 The densely packed nucleic acid shell effectively protects the oligonucleotides from nuclease degradation, thereby significantly extending their circulation half-life in complex biological environments. Meanwhile, the inorganic nanoparticle core (such as gold nanoparticles) provides robust structural support, ensuring the overall integrity and reproducibility of the construct. Second, the most striking feature of SNAs lies in their remarkably high cellular uptake efficiency.179 Unlike conventional linear nucleic acids that require transfection reagents to enter cells, SNAs can be actively and efficiently internalized by a wide range of cell types—including traditionally hard-to-transfect primary cells—via clathrin-mediated endocytosis, thus overcoming one of the major barriers in nucleic acid drug delivery. Moreover, the high-density oligonucleotide shell of SNAs generates a pronounced multivalent effect, which not only enhances their hybridization affinity toward complementary sequences but also provides a versatile platform for molecular functionalization.180 By co-conjugating different types of functional nucleic acids (e.g., siRNA or aptamers) or chemical moieties on the same nanoparticle surface, SNAs can achieve targeted delivery, synergistic therapy, and stimuli-responsive behavior, offering unprecedented freedom in molecular design and programmability in biological function.
Building upon these advantages, SNAs have demonstrated significant value in the design and delivery of nucleic acid therapeutics. In terms of delivery, SNAs can be efficiently internalized by a wide variety of cell types without the need for transfection agents, thereby avoiding the cytotoxicity and immunogenicity commonly associated with traditional nonviral vectors. For example, Jie Li et al. proposed an interface engineering strategy based on a tetrahedral DNA framework (tDF) to construct a novel DNA framework spherical nucleic acid (tDF-SNA).181 In this design, siRNA-loaded tDFs were precisely anchored onto gold nanoparticle surfaces through Au–S bonds, forming corona-like spike structures with flexible conformations around the nanoparticle core. This architecture led to a 1–2 order of magnitude increase in siRNA delivery efficiency and approximately a twofold enhancement in specific gene-silencing activity (Fig. 8B). In addition, SNAs with lipid or polymeric cores (such as liposomal SNAs and polymer-core SNAs) have been extensively employed for the delivery of mRNA and ASOs, exhibiting improved serum stability and prolonged in vivo circulation times.182 Furthermore, the three-dimensional spherical topology of SNAs confers pronounced multivalency and cooperative recognition capabilities.183 By arranging multiple functional oligonucleotides with precise spatial control on a single nanoparticle surface, SNAs can simultaneously mediate multivalent molecular recognition and synergistic regulation within a unified nanoscale platform. On this basis, Wang et al. developed an intelligent SNA system in which two antisense oligonucleotides complementary to the oncogenic miRNAs miR-21 and miR-155 were covalently grafted onto gold nanoparticles. In addition, the chemotherapeutic drug DOX and a photosensitizer were hybridized onto the antisense strands. This multifunctional SNA could simultaneously capture target miRNAs and release both the photosensitizer and DOX in a controlled manner, thereby achieving combined gene, photodynamic, and chemotherapeutic effects within a single nanosystem (Fig. 8C).184 At the clinical translation level, SNA technology has begun to move toward practical application. The SNA platform developed by Exicure Inc. (including AST-008) has been employed as a Toll-like receptor 9 (TLR9) agonist for immunotherapy and is currently under clinical investigation for the treatment of melanoma and breast cancer.185 Moreover, the SNA-based drug XCUR17, designed for psoriasis therapy, demonstrated favorable safety and significant gene-silencing efficacy in a Phase I clinical trial.186 Collectively, these studies highlight that SNAs not only exhibit superior performance in nucleic acid delivery and tissue penetration, but also hold broad potential in cancer therapy, immunomodulation, and the treatment of inflammatory diseases.
Compared with traditional nanocarriers such as liposomes, polymeric nanoparticles, and inorganic materials, DNA origami shows significant advantages.192 First, DNA origami possesses a high degree of programmability and predictable architecture.193 Benefiting from the Watson–Crick base-pairing principle, researchers can precisely control the position and pairing of each nucleotide through computer-aided design, thereby achieving customizable construction of nanoscale structures with defined morphology, size, and topology. This unprecedented level of structural accuracy allows DNA origami to be designed in various forms—such as rod-like, tubular, box-shaped, cage-like, or even dynamic architectures capable of opening, closing, or conformational switching—to meet diverse requirements for drug loading and targeted delivery. Second, DNA origami demonstrates excellent spatial addressability and functional modularity.194 Each DNA strand within the structure can be regarded as a distinct addressable site. By extending the staple strands or introducing chemical modifications, functional entities such as proteins, peptides, chemotherapeutic drugs, nucleic acid aptamers, and siRNAs can be anchored at predetermined positions with defined copy numbers, geometric arrangements, and inter-ligand spacings. This precise spatial control provides an ideal platform for investigating multivalent interactions and constructing multifunctional or stimuli-responsive therapeutic systems. Finally, DNA origami offers notable advantages in biocompatibility and biodegradability.195 As a nanostructure composed of natural biomacromolecules, its framework can be enzymatically degraded in vivo into nontoxic nucleotide byproducts, minimizing biosafety concerns. Compared with most inorganic or polymeric delivery vehicles, DNA origami achieves precise molecular delivery while offering superior intrinsic safety and greater potential for clinical translation.
Building upon these advantages, DNA origami has gradually evolved into a precisely designable and highly programmable platform for nucleic acid therapeutics, showing great potential in drug delivery, targeted recognition, and multimodal combination therapy.196 In programmable drug delivery, DNA origami can serve as an accurate carrier for molecular loading and controlled release. By spatially positioning nucleic acid sequences, it enables efficient transport and programmable release of therapeutic payloads. Wang et al. employed DNA origami technology to construct a functionalized DNA nanodevice in which siRNA was encapsulated within the inner cavity, while the chemotherapeutic drug DOX was intercalated into the DNA duplexes.197 The incorporation of disulfide linkages into the framework allowed GSH-triggered release of siRNA under the reductive environment of tumor cells, achieving precise gene silencing of oncogenic targets and significantly suppressing cancer progression (Fig. 8D). The spatially programmable nature of DNA origami allows it to achieve multivalent recognition on the nanoscale. By precisely tuning the distance, density, and orientation of recognition motifs, researchers can systematically investigate how receptor clustering, signal transduction, and immune recognition depend on spatial organization. Zhang et al. constructed DNA-origami-based arrays displaying receptor-binding domains (RBDs) of SARS-CoV-2 with defined valencies and spacings, revealing how nanoscale ligand organization governs viral infection efficiency and immune activation mechanisms (Fig. 8E).198 Hu et al. further designed a series of tunable multivalent aptamer-modified DNA nanostructures, in which the aptamer type, valency, binding pattern, and origami geometry were adjusted to modulate tumor-targeting selectivity.199 Tubular origami structures were employed to deliver prodrugs into tumor cells, while sheet-like structures facilitated specific interactions between macrophages and tumor cells, thereby promoting immune clearance. These studies collectively highlight the spatial programmability of DNA origami in achieving multivalent recognition and immune modulation (Fig. 8F). In addition, DNA origami provides a modular platform for combination therapy. Xu et al. designed an octahedral DNA origami framework (OctDOFs) capable of co-loading siRNA, the chemotherapeutic agent DOX, and gold nanorods as photothermal agents, thereby integrating gene silencing, chemotherapy, and photothermal therapy into a single multimodal nanotherapeutic system (Fig. 8G).200
Overall, the emergence of nucleic acid nanostructures—such as spherical nucleic acids (SNAs) and DNA origami—has propelled nucleic acid therapeutics from sequence-level optimization toward structural-level innovation, providing new strategies for precise delivery and intelligent therapy.
3. Challenge in nucleic acid development
3.1. Delivery issues
At present, nucleic acid therapeutics are predominantly administered via parenteral routes, with intravenous and subcutaneous injections being the most common, while oral formulations remain in the early exploratory stage.201The vascular endothelium—particularly specialized barriers such as the blood–brain barrier—further limits drug penetration into target tissues.203 Only under pathological conditions (e.g., tumors or inflammation) where vascular permeability is enhanced can the enhanced permeability and retention (EPR) effect facilitate drug accumulation at diseased sites. Even after traversing endothelial barriers, nucleic acid therapeutics face additional onstacles within the extracellular matrix, such as fibrosis and elevated interstitial pressure, which hinder their diffusion and effective distribution to target cells. Upon reaching target tissues, nucleic acid therapeutics must further enter target cells and, in some cases, reach specific subcellular compartments. However, the negative charges on nucleic acids result in electrostatic repulsion from the similarly negatively charged cell membrane,204 which hinders their transmembrane transport. Cellular uptake usually relies on cationic carrier-mediated endocytosis. Once internalized, nucleic acids typically first localize to early endosomes and are subsequently trafficked to late endosomes and lysosomes, nucleic acids are initially localized to early endosomes and eventually trafficked to late endosomes or lysosomes, where the acidic environment and nuclease increase their susceptibility to degradation.205 Efficient endosomal escape is therefore a central challenge in nucleic acid delivery, with strategies focusing on pH-responsive materials or membrane-disruptive mechanisms to enable timely release into the cytoplasm. Additionally, for nucleic acid therapeutics requiring nuclear entry, crossing the nuclear envelope is particularly challenging in non-dividing cells, often necessitating specific nuclear localization signals or carrier systems for efficient nuclear delivery.
In summary, the selection of an appropriate administration route for nucleic acid therapeutics should comprehensively consider the target organ, physicochemical properties of the molecule, and desired pharmacokinetic profile. Intravenous delivery is challenged by systemic barriers and limited cellular uptake, whereas subcutaneous and intramuscular routes are constrained by local absorption and systemic transport. In contrast, the key determinants for intrathecal and local injections lie in achieving efficient distribution, prolonged retention, and effective cellular transfection within the target region.
3.2. Stability and safety
To improve stability, several strategies have been developed: (1) chemical modifications: phosphorothioate substitution, 2′-O-methylation, and incorporation of LNA structures significantly improve stability.225 (2) Terminal capping: modifying the 3′ and 5′ ends to block nuclease recognition site.226 (3) Conjugation with delivery systems: encapsulation in lipid nanoparticles,227 polymers,228 or proteins229 can prolong circulation time, reduce degradation, and enhance bioavailability.
3.3. Cost and scalability
Following synthesis, purification and quality control processes emerge as the primary bottlenecks in cost reduction and scalability. Techniques such as high-performance liquid chromatography (HPLC), ultrafiltration, and gel electrophoresis are commonly employed to remove impurities, such as short fragments, unmodified strands, and by-products. However, these methods demand expensive equipment, high reagent consumption, and time-intensive operation, which hinder large-scale production and drive-up costs. For instance, in the production of Onpattro (patisiran), HPLC purification was a critical limiting factor that restricted capacity and significantly increased costs. Moreover, stringent quality control is required throughout the process, including LC-MS, capillary electrophoresis (CE), and qPCR to ensure sequence integrity and correct chemical modifications. For mRNA vaccines, additional tests are required to verify capping efficiency and lipid nanoparticle (LNP) encapsulation, further extending production timelines and increasing per-unit cost. While essential for ensuring safety and efficacy, these rigorous processes pose major challenges to scaling production and achieving cost reductions during commercialization.
Large-scale LNP manufacturing typically relies on continuous-flow microfluidics, where organic and aqueous phases are mixed to produce uniform nanoparticles. While this method offers superior reproducibility, particle size control, and encapsulation efficiency compared with traditional bulk mixing, it entails high capital investment and maintenance costs, as well as stringent requirements for operational environments (e.g., sterile conditions, precise temperature control). Moreover, technology transfer between production lines necessitates extensive validation batches, further inflating the fixed costs of early commercialization.
An illustrative case of the scale-up challenge occurred with of the production of COVID-19 vaccines. Despite having efficient mRNA synthesis pipelines, companies like Pfizer/BioNTech and Moderna faced significant bottlenecks in LNP encapsulation capacity. Early in BNT162b2 production, BioNTech had to rely on external partners, such as Acuitas Therapeutics and Polymun Scientific, for LNP formulation, as internal production capacity was insufficient. This resulted in delays due to equipment limitations, high demands for batch-to-batch consistency, and lengthy validation cycles.
3.4. Long-term efficacy and adverse effect monitoring
The durability of efficacy and long-term safety are central factors in determining the clinical value of nucleic acid therapeutics. Over time, repeated dosing may lead to reduced efficacy, driven by several mechanisms. The immune system can generate specific antibodies against delivery systems, such as lipid nanoparticles or PEGylated components, leading faster drug clearance and altered tissue distribution. Additionally, target genes may evade therapeutic inhibition through mutations, production of alternative splice variants, or activation of compensatory signaling pathways.241 Furthermore, intracellular delivery and release efficiency may decline due to factors such as saturation of RISC,242 reduced endocytosis, or impaired endosomal escape. In addition to efficacy concers, long-term administration also raises concerns regarding chronic toxicities, including accumulation of drugs or delivery materials in organs such as the liver and kidney, persistent low-grade inflammatory responses, and potential off-target effects or genomic instability associated with gene-editing therapies.To comprehensively evaluate these risks, a systematic and standardized long-term follow-up framework is urgently needed to enable dynamic monitoring of both efficacy and safety. Such a framework should span from baseline to multi-year assessments, incorporating not only disease-specific clinical endpoints and biomarker changes but also pharmacokinetic/pharmacodynamic data. Longitudinal monitoring should include immunological indices (e.g., anti-drug antibody titers, cytokine profiles), organ function (e.g., hepatic and renal function, coagulation markers), and molecular-level changes (e.g., target gene expression, alternative splicing patterns, potential off-target events).243 In parallel, integration of real-world registry studies with remote digital health monitoring can provide continuous insights into patients' quality of life and functional status, with pre-defined alert thresholds enabling timely intervention. Such a long-term monitoring framework will not only safeguard the therapeutic efficacy and safety of nucleic acid drugs across the treatment course but also provide critical evidence for the rational design and optimization of next-generation nucleic acid therapies.
4. Future perspectives and opportunities
4.1. CRISPR and gene editing therapies
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and its derivative gene-editing systems (such as CRISPR–Cas9, Cas12a, and Cas13) have revolutionized nucleic acid therapeutics by enabling precise genome editing.244 Unlike conventional strategies that rely on exogenous nucleic acids for supplementation or inhibition, CRISPR allows for direct modifications at the genomic level—including gene knockout, knock-in, and base substitution—thereby addressing pathogenic mutations at their source. Gene-editing therapies hold the potential to achieve long-lasting or even permanent therapeutic effects with a single administration and can target previously considered “undruggable”. Prominent successes include the restoration of function in DMD models using Cas9, and the capacity of Cas13 to specifically target viral RNA transcripts.245Despite these advances, clinical application of CRISPR faces significant challenges, particularly in delivery efficiency and safety. Currently, such as adeno-associated virus (AAV) vectors and lipid nanoparticles (LNPs) are the primary delivery systems for CRISPR components (Cas proteins and their guide RNAs).246
However, these approaches require a delicate balance between delivery efficiency, immunogenicity, and durability. While AAV-mediated gene editing has advanced into clinical trials for ocular and liver diseases, concerns regarding vector integration and immune response remain. Non-viral strategies, such as LNPs carrying mRNA encoding Cas proteins,247 can mitigate integration risks and allow repeat dosing, but issues related to tissue specificity, editing efficiency, and off-target effects continue to pose significant barriers
Looking ahead, CRISPR-based therapies are expected to extend beyond monogenic disorders into complex polygenic diseases, viral infections, and cancer immunomodulation. Emerging innovations, such as high-fidelity Cas variants (e.g., Cas9-HF, eSpCas9),248 base editors (BEs), and prime editors (PEs), are improving precision and minimizing off-target risks, thereby improving editing precision and safety.249 The integration of AI-driven guide RNA design, along with inducible systems for spatiotemporal control of gene editing, will further enhance the predictability and precision of these therapies. Together with advancements in delivery systems and monitoring technologies, CRISPR-based gene editing is poised to significantly impact precision and personalized medicine
4.2. Synthetic biology and nanotechnology
Synthetic biology offers new avenues for advancing nucleic acid therapeutics by incorporating programmable features that enhance efficacy and safety. Tools, such as synthetic promoters, toehold switches, riboswitches, CRISPRi/a systems, and base or prime editors, can be engineered to control gene expression or editing activity in response to specific signals, cell types, or timeframes.250 This precision enables logic-gated regulation (e.g., AND/NOT functions), which reduces off-target risks and systemic side effects. For example, self-amplifying RNAs (saRNAs) and circular RNAs (circRNAs) can amplify therapeutic effects and extend expression duration under limited doses.251 In addition, “safety switches” (kill-switches) and inducible termination systems (e.g., drug-inducible Cas proteins or molecular degraders) allow rapid suspension of activity in the event of adverse reactions. In terms of targeting strategies, aptamers or peptide ligands can recognize specific receptors on cell surfaces (e.g., GalNAc for hepatocyte targeting or tumor-specific aptamers),252 thereby directing biodistribution and cellular selectivity at the earliest stages of drug delivery. This “navigation” function shifts the decision point of drug specificity to the initial delivery step, enabling integrated optimization across delivery, cellular uptake, and functional activity, which enhances therapeutic selectivity while minimizing off-target toxicity.Nanotechnology provides a crucial role in enableing the delivery of these programmable strategies. Lipid nanoparticles (LNPs) are widely used due to their ability to efficiently encapsulate and deliver nucleic acids while ensuring effective endosomal escape. Other carriers, including polymeric and inorganic carriers, utilize stimulus-responsive designs (e.g., pH, redox, or enzymatic triggers) for controlled release in specific microenvironments. DNA origami and nucleic acid nanostructures provide precise multivalent delivery,253 enabling the co-delivery of genome-editing tools and repair templates. Biomimetic carriers, such as exosomes, enhance immune compatibility and circulation time, promoting more effective and sustained therapeutic delivery. From a translational perspective, continuous-flow microfluidic assembly and modularized quality-control systems can ensure particle size uniformity and batch-to-batch stability while reducing per-dose costs and shortening release cycles. Together, synthetic biology determines “when, where, and how” therapeutic functions are exerted, while nanotechnology ensures they are “delivered and released efficiently.” The synergy of these two disciplines thus establishes an integrated pathway for nucleic acid therapeutics, spanning precise action, safety regulation, and scalable manufacturing.
4.3. Combination therapy
The integration of nucleic acid drugs with small molecules or antibodies offers synergistic therapeutic potential, particularly in enhancing pathway complementarity and reversing resistance mechanisms. For example, siRNA or ASO can downregulate key drivers or resistance factors (e.g., KRAS adaptors, BCL2, ABCB1/P-gp), 254–256 thereby resensitizing tumors to targeted therapies or chemotherapy. In the immunotherapy, mRNA/siRNA modulation of immune axes such as PD-L1 and IL-12 can amplify both the depth and durability of responses to PD-1/PD-L1 inhibitors (e.g., combining PD-L1 siRNA or IL-12 mRNA with anti-PD-1 therapy). Such synergies are often sequence-dependent, where nucleic acid drugs act as a “pre-conditioning” step to suppress escape or resistance pathways before chemotherapy or targeted therapy. In terms of delivery, co-encapsulation within a single carrier (ensuring co-delivery into the same cells) and separate administration (reducing formulation complexity) are viable options. Real-time decision-making can be guided by disease biomarkers and pharmacokinetic/pharmacodynamic readouts, such as target protein downregulation and inhibition of downstream signalling.As activators of inert small-molecule prodrugs, nucleic acid drugs can function as spatiotemporally controllable “molecular switches.” A representative strategy is gene- or mRNA-directed enzyme prodrug therapy (GDEPT/mRNA-DEPT), in which DNA or mRNA encoding an activating enzyme is delivered to ensure enzyme expression is restricted to the lesion site,257 thereby converting systemically administered prodrugs into cytotoxic metabolites with high selectivity and a “bystander effect.” Classical enzyme-prodrug pairs include HSV-TK/ganciclovir,258 cytosine deaminase (CD)/5-fluorocytosine,259 and nitroreductase/CB1954.260 Another approach employs nucleic acid nanostructures or aptamers as “locks,” which are “unlocked” upon recognition of tumor biomarkers (e.g., specific receptors or miRNAs) to release embedded chemotherapeutics (such as DOX). Similarly, CRISPRa and switch-type riboswitches can be designed to induce enzyme expression under tumor-specific promoters.261 The shared advantage of these strategies lies in restricting pharmacological activity to the lesion site, thereby markedly expanding the therapeutic window. Nonetheless, clinical translation remains hindered by several bottlenecks, including spatial co-localization and expression heterogeneity between the activation system and prodrug, immunogenicity during delivery, and challenges of manufacturing consistency across batches. For successful development, it is imperative to establish key clinical endpoints, such as tissue-specific expression levels, the ratio of prodrug to active drug in vivo, and long-term safety readouts (immunological and organ function). Concurrently, replicable dosing regimens and companion diagnostic frameworks must be implemented to enable genuine therapeutic synergy and toxicity reduction.
4.4. Construction of novel delivery vectors
The design of novel nucleic acid delivery vectors has evolved from focusing on single-material optimization to a more holistic, multidimensional approach that integrates material selection, structural design, functional module integration, and large-scale manufacturing. Ionizable lipids are fine-tuned for optimal loading efficiency and endosomal escape, while degradable polyesters and poly (β-amino ester) polymers help mitigate safety concerns related to accumulation.262 Amphiphilic peptides and protein nanocages provide greater structural programmability and multivalent ligand display capabilities; while nucleic acid nanostructures such as DNA origami enable precise spatial configuration control at the nanoscale.263From a structural perspective, core–shell architectures, multilamellar vesicles, or dendrimeric frameworks can simultaneously enhance stability and loading capacity. Surface modification with hydrophilic polymers or alternative hydrophobic coronas (e.g., low-immunogenic PEG substitutes) prolongs circulation time and improves immune tolerance. At the functional module level, incorporation of endosomal escape elements, stimuli-responsive release mechanisms (pH, enzymatic, reductive, or light triggers), and SORT lipids enables spatiotemporally precise delivery of nucleic acid therapeutics. Finally, at the manufacturing level, continuous-flow microfluidics and modular production platforms markedly improve batch-to-batch consistency while meeting GMP requirements, thereby laying the foundation for large-scale clinical-grade manufacturing.
Hybrid multi-carrier strategies, by integrating the advantages of distinct delivery platforms while offsetting their respective limitations, hold great promise for further expanding the therapeutic window of nucleic acid drugs. For instance, lipid-polymer nanoparticles (LPNs) employ polymeric cores to achieve enhanced mechanical stability and higher loading capacity,264 while relying on lipid shells to ensure favorable biocompatibility and efficient endosomal escape. Lipid nanoparticles cloaked with exosomes or cellular membranes markedly improve immune evasion and confer tissue- or organ-specific targeting capabilities. Virus-like particle (VLP)–LNP composite systems enable the co-delivery or sequential release of Cas mRNA, sgRNA, and repair templates, thereby fulfilling the demands of complex genome-editing applications.265 Moreover, combining these carriers with microneedles, injectable hydrogels, or physical triggering modalities (such as ultrasound-microbubble or magnetically responsive systems) allows for localized, efficient, and controllable drug release. It is important to note, however, that hybrid multi-component systems often encounter greater challenges in clinical translation, including formulation stability, immunogenicity, and regulatory compliance. Thus, the principle of “minimal sufficient complexity” should be followed—favoring degradable, chemically well-defined, and modular platforms with scalable manufacturing potential—while their clinical advantages must be validated through systematic pharmacodynamic and safety evaluations.
4.5. Precision-controlled drug release
Achieving precise release of nucleic acid therapeutics requires programming the “recognition–delivery–release” cascade and regulating it across both spatial and temporal dimensions. At the spatial level, receptor–ligand strategies (e.g., GalNAc-mediated hepatocyte targeting, aptamer/peptide- or antibody-guided tumor selectivity)266–268 and organ-selective lipids enable pre-selection of specific tissues and cell types.269 At the subcellular level, optimization of ionizable lipid pKa and incorporation of endosomal escape modules allow the capture of the critical “time window” from endocytosis to cytoplasmic release. At the temporal level, stimuli-responsive mechanisms (pH, redox, enzymatic cleavage, ROS) and exogenous triggers (light, magnetic fields, ultrasound) can finely control the release rate and initiation timing. For expression-based cargos, synthetic biology tools such as riboswitches, toehold switches, and CRISPRa/i induction systems, in combination with molecular logic gates (AND/NOT), ensure that pharmacological activity is activated only when specific signals are met, thereby minimizing off-target effects and systemic toxicity. Depending on therapeutic needs, long-lasting interventions may exploit circular RNA or self-amplifying RNA (saRNA) to extend duration of action, whereas scenarios requiring intermittent stimulation can leverage microneedles or injectable hydrogels as local depots to achieve pulsatile or sequential release.Multistage delivery and closed-loop control represent critical pathways for enhancing clinical controllability. On the one hand, coupling the three key elements of “signal sensing-conditional decision-responsive release” (e.g., using disease-associated miRNAs or proteins as triggering inputs and carrier disassembly or transcriptional initiation as outputs) enables the construction of adaptive feedback release systems. On the other hand, complex therapeutic regimens can be coordinated through co-delivery within the same carrier or sequential administration in separate formulations—for example, the staged release of Cas mRNA, sgRNA, and repair templates, or the combined application of nucleic acids with small molecules.270,271 These strategies can be dynamically calibrated by pharmacokinetic/pharmacodynamic (PK/PD) readouts, including tissue exposure levels, target inhibition rates, and cytokine profiles, to refine dosing rhythms in real time. In parallel, manufacturability and regulatory compliance must be considered. Continuous-flow microfluidics and modular CMC frameworks provide robust platforms to predefine critical quality attributes (e.g., particle size and polydispersity, encapsulation efficiency, leakage rate, activation threshold, and release kinetics). Quality-by-Design (QbD) methodologies further ensure batch-to-batch consistency and reproducibility of exogenous triggers. Through such programmed, spatiotemporally integrated strategies, nucleic acid therapeutics may achieve higher therapeutic indices and more predictable clinical outcomes while maintaining safety.
Author contributions
All authors contributed to the writing and revision of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Data availability
No primary research results, software or code have been included, and no new data were generated or analyzed as a part of this perspective.Acknowledgements
This work was funded by the National Key Research and Development Program of China (2021YFA0910000).Notes and references
- C. Bustamante, W. Cheng and Y. X. Mejia, Revisiting the central dogma one molecule at a time, Cell, 2011, 144, 480–497 Search PubMed.
- R. L. P. Adams, J. T. Knowler and D. P. Leader, The biochemistry of the nucleic acids, Springer Science & Business Media, 2013 Search PubMed.
- Nucleic Acids in Chemistry and Biology, ed. G. M. Blackburn, M. J. Gait, D. Loakes and D. Williams, Royal Society of Chemistry, 2006 Search PubMed.
- X. Bian, L. Zhou, Z. Luo, G. Liu, Z. Hang, H. Li, F. Li and Y. Wen, Emerging delivery systems for enabling precision nucleic acid therapeutics, ACS Nano, 2025, 19, 4039–4083 Search PubMed.
- J. B. Opalinska and A. M. Gewirtz, Nucleic-acid therapeutics: basic principles and recent applications, Nat. Rev. Drug Discovery, 2002, 1, 503–514 CrossRef CAS PubMed.
- J. D. Watson and F. H. Crick, Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid, Nature, 1953, 171, 737–738 Search PubMed.
- N. N. Danial and S. J. Korsmeyer, Cell death: critical control points, Cell, 2004, 116, 205–219 Search PubMed.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, DNA-based, method for rationally assembling nanoparticles into macroscopic materials/Ch, Nature, 1996, 382, 607–609 Search PubMed.
- P. W. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 Search PubMed.
- K. Garber, Alnylam launches era of RNAi drugs, Nat. Biotechnol., 2018, 36, 777–778 CrossRef CAS PubMed.
- P. M. Cannon and H.-P. Kiem, The genome-editing decade, Mol. Ther., 2021, 29, 3093–3094 CrossRef CAS PubMed.
- N. Mandal, Nobel Prize in physiology or medicine 2023: development of mRNA therapeutics that paved the path of formulation of COVID-19 vaccine, J. Hem. Allied Sci., 2024, 3, 81–87 Search PubMed.
- D. C. Henshall, MicroRNAs, Cambridge University Press, 2024 Search PubMed.
- X. Cheng, Y. Wang, T. Zhang, Y. Cao, H. Cheng, Y. Du, X. Ge, B. Ren, J. Lu and L. Li, Emergence of Small Nucleic Acids as Drugs in Precision Medicine, Clin. Pharmacol. Ther., 2025, 118, 308–323 Search PubMed.
- E. S. Smith, E. Whitty, B. Yoo, A. Moore, L. F. Sempere and Z. Medarova, Clinical applications of short non-coding RNA-based therapies in the era of precision medicine, Cancers, 2022, 14, 1588 Search PubMed.
- A. Sadri, Is target-based drug discovery efficient? Discovery and “off-target” mechanisms of all drugs, J. Med. Chem., 2023, 66, 12651–12677 CrossRef CAS PubMed.
- D. Brown, Unfinished business: target-based drug discovery, Drug discovery today, 2007, 12, 1007–1012 CrossRef CAS PubMed.
- C. G. Jackson, in Reducing Drug Attrition, Springer, 2014, vol. 11, pp. 1–72 Search PubMed.
- H. Gharwan and H. Groninger, Kinase inhibitors and monoclonal antibodies in oncology: clinical implications, Nat. Rev. Clin. Oncol., 2016, 13, 209–227 Search PubMed.
- J. L. Adams, J. Smothers, R. Srinivasan and A. Hoos, Big opportunities for small molecules in immuno-oncology, Nat. Rev. Drug Discovery, 2015, 14, 603–622 CrossRef CAS PubMed.
- G. Rodgers, C. Austin, J. Anderson, A. Pawlyk, C. Colvis, R. Margolis and J. Baker, Glimmers in illuminating the druggable genome, Nat. Rev. Drug Discovery, 2018, 17, 301–302 Search PubMed.
- D. Kandari and R. Bhatnagar, Antibody engineering and its therapeutic applications, Int. Rev. Immunol., 2023, 42, 156–183 Search PubMed.
- L. G. Presta, Selection, design, and engineering of therapeutic antibodies, J. Allergy Clin. Immunol., 2005, 116, 731–736 CrossRef CAS PubMed.
- W. Jiskoot, A. Hawe, T. Menzen, D. B. Volkin and D. J. Crommelin, Ongoing challenges to develop high concentration monoclonal antibody-based formulations for subcutaneous administration: quo vadis?, J. Pharm. Sci., 2022, 111, 861–867 Search PubMed.
- L. M. Alvarez-Salas, Nucleic acids as therapeutic agents, Curr. Top. Med. Chem., 2008, 8, 1379–1404 CrossRef CAS PubMed.
- C. Chen, Z. Yang and X. Tang, Chemical modifications of nucleic acid drugs and their delivery systems for gene-based therapy, Med. Res. Rev., 2018, 38, 829–869 CrossRef PubMed.
- Y. Zhao, R. Shu and J. Liu, The development and improvement of ribonucleic acid therapy strategies, Mol. Ther. Nucleic Acids, 2021, 26, 997–1013 CrossRef CAS PubMed.
- A. Sang, S. Zhuo, A. Bochanis, J. E. Manautou, R. Bahal, X.-b. Zhong and T. P. Rasmussen, Mechanisms of action of the US food and drug administration-approved antisense oligonucleotide drugs, BioDrugs, 2024, 38, 511–526 CrossRef CAS PubMed.
- Z. Dominski and R. Kole, Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 8673–8677 CrossRef CAS PubMed.
- F. J. Raal, R. D. Santos, D. J. Blom, A. D. Marais, M.-J. Charng, W. C. Cromwell, R. H. Lachmann, D. Gaudet, J. L. Tan, S. Chasan-Taber, D. L. Tribble, J. D. Flaim and S. T. Crooke, Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial, Lancet, 2010, 375, 998–1006 CrossRef CAS PubMed.
- M. E. Visser, J. L. Witztum, E. S. G. Stroes and J. J. P. Kastelein, Antisense oligonucleotides for the treatment of dyslipidaemia, Eur. Heart J., 2012, 33, 1451–1458 CrossRef CAS PubMed.
- X.-H. Liang, H. Sun, J. G. Nichols and S. T. Crooke, RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus, Mol. Ther., 2017, 25, 2075–2092 CrossRef CAS PubMed.
- B. P. Monia, E. A. Lesnik, C. Gonzalez, W. F. Lima, D. McGee, C. J. Guinosso, A. M. Kawasaki, P. D. Cook and S. M. Freier, Evaluation of 2'-modified oligonucleotides containing 2'-deoxy gaps as antisense inhibitors of gene expression, J. Biol. Chem., 1993, 268, 14514–14522 CrossRef CAS PubMed.
- S. Emmrich, W. Wang, K. John, W. Li and B. M. Pützer, Antisense gapmers selectively suppress individual oncogenic p73 splice isoforms and inhibit tumor growth in vivo, Mol. Cancer, 2009, 8, 61 CrossRef PubMed.
- M. A. Havens and M. L. Hastings, Splice-switching antisense oligonucleotides as therapeutic drugs, Nucleic Acids Res., 2016, 44, 6549–6563 CrossRef PubMed.
- N. N. Singh, M. N. Lawler, E. W. Ottesen, D. Upreti, J. R. Kaczynski and R. N. Singh, An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy, Nucleic Acids Res., 2013, 41, 8144–8165 CrossRef CAS PubMed.
- R. Peralta, A. Low, A. Kim, S. Murray, S. Guo, S. Freier, T. M. Townes and G. Hung, Targeting BCL11A and KLF1 For the Treatment of Sickle Cell Disease and β-Thalassemia In Vitro using Antisense Oligonucleotides, Blood, 2013, 122, 1022 CrossRef.
- K. Yang, W. Zhang, L. Zhong, Y. Xiao, S. Sahoo, M. Fassan, K. Zeng, P. Magee, M. Garofalo and L. Shi, Long non-coding RNA HIF1A-As2 and MYC form a double-positive feedback loop to promote cell proliferation and metastasis in KRAS-driven non-small cell lung cancer, Cell Death Differ., 2023, 30, 1533–1549 CrossRef CAS PubMed.
- D. Collotta, I. Bertocchi, E. Chiapello and M. Collino, Antisense oligonucleotides: a novel Frontier in pharmacological strategy, Front. Pharmacol, 2023, 14, 1304342 CrossRef CAS PubMed.
- L. Cartegni and A. R. Krainer, Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1, Nat. Genet., 2002, 30, 377–384 CrossRef CAS PubMed.
- S. R. Lim and K. J. Hertel, Modulation of Survival Motor Neuron Pre-mRNA Splicing by Inhibition of Alternative 3′ Splice Site Pairing, J. Biol. Chem., 2001, 276, 45476–45483 CrossRef CAS PubMed.
- A. Khvorova, Modulation of DNA transcription: The future of ASO therapeutics?, Cell, 2022, 185, 2011–2013 CrossRef CAS PubMed.
- M. Haché, K. J. Swoboda, N. Sethna, A. Farrow-Gillespie, A. Khandji, S. Xia and K. M. Bishop, Intrathecal Injections in Children with Spinal Muscular Atrophy, J. Child Neurol., 2016, 31, 899–906 CrossRef PubMed.
- D. R. Corey, Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy, Nat. Neurosci., 2017, 20, 497–499 CrossRef CAS PubMed.
- G. S. Thomas, W. C. Cromwell, S. Ali, W. Chin, J. D. Flaim and M. Davidson, Mipomersen, an Apolipoprotein B Synthesis Inhibitor, Reduces Atherogenic Lipoproteins in Patients With Severe Hypercholesterolemia at High Cardiovascular Risk: A Randomized, Double-Blind, Placebo-Controlled Trial, J. Am. Coll. Cardiol., 2013, 62, 2178–2184 CrossRef CAS PubMed.
- F. Akdim, E. S. G. Stroes, E. J. G. Sijbrands, D. L. Tribble, M. D. Trip, J. W. Jukema, J. D. Flaim, J. Su, R. Yu, B. F. Baker, M. K. Wedel and J. J. P. Kastelein, Efficacy and Safety of Mipomersen, an Antisense Inhibitor of Apolipoprotein B, in Hypercholesterolemic Subjects Receiving Stable Statin Therapy, J. Am. Coll. Cardiol., 2010, 55, 1611–1618 CrossRef CAS PubMed.
- A. Pirillo, G. Norata and A. Catapano, Advances in Hypercholesterolemia, Compr. Med. Chem. III, 2017, 663–693 CAS.
- D. Duan, N. Goemans, S. i. Takeda, E. Mercuri and A. Aartsma-Rus, Duchenne muscular dystrophy, Nat. Rev. Dis. Primers, 2021, 7, 13 CrossRef PubMed.
- E. M. McNally and E. J. Wyatt, Mutation-based therapy for duchenne muscular dystrophy: Antisense treatment arrives in the clinic, Circulation, 2017, 136, 979–981 CrossRef PubMed.
- J. R. Mendell, N. Goemans, L. P. Lowes, L. N. Alfano, K. Berry, J. Shao, E. M. Kaye, E. Mercuri, F. T. E. S. Group and T. F. D. I. Network, Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy, Ann. Neurol., 2016, 79, 257–271 CrossRef CAS PubMed.
- C. Terada, S. Kawamoto, A. Yamayoshi and T. Yamamoto, Chemistry of therapeutic oligonucleotides that drives interactions with biomolecules, Pharmaceutics, 2022, 14, 2647 CrossRef CAS PubMed.
- N. Iwamoto, D. C. D. Butler, N. Svrzikapa, S. Mohapatra, I. Zlatev, D. W. Y. Sah, Meena, S. M. Standley, G. Lu, L. H. Apponi, M. Frank-Kamenetsky, J. J. Zhang, C. Vargeese and G. L. Verdine, Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides, Nat. Biotechnol., 2017, 35, 845–851 CrossRef CAS PubMed.
- M. K. Bijsterbosch, E. T. Rump, R. L. A. D. Vrueh, R. Dorland, R. van Veghel, K. L. Tivel, E. A. L. Biessen, T. J. C. van Berkel and M. Manoharan, Modulation of plasma protein binding and in vivo liver cell uptake of phosphorothioate oligodeoxynucleotides by cholesterol conjugation, Nucleic Acids Res., 2000, 28, 2717–2725 CrossRef CAS PubMed.
- D. Engelhardt, P. Nordberg, L. Knerr and L. R. Malins, Accessing Therapeutically-Relevant Multifunctional Antisense Oligonucleotide Conjugates Using Native Chemical Ligation, Angew. Chem., Int. Ed., 2024, 63, e202409440 CrossRef CAS PubMed.
- S. Obika, D. Nanbu, Y. Hari, J.-i. Andoh, K.-i. Morio, T. Doi and T. Imanishi, Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides, Tetrahedron Lett., 1998, 39, 5401–5404 CrossRef CAS.
- T. Yoshida, K. Morihiro, Y. Naito, A. Mikami, Y. Kasahara, T. Inoue and S. Obika, Identification of nucleobase chemical modifications that reduce the hepatotoxicity of gapmer antisense oligonucleotides, Nucleic Acids Res., 2022, 50, 7224–7234 CrossRef CAS PubMed.
- A. M. Kawasaki, M. D. Casper, S. M. Freier, E. A. Lesnik, M. C. Zounes, L. L. Cummins, C. Gonzalez and P. D. Cook, Uniformly modified2'-deoxy-2'-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets, J. Med. Chem., 1993, 36, 831–841 Search PubMed.
- F. Ramírez-Cortés and P. Ménová, Hepatocyte targeting via the asialoglycoprotein receptor, RSC Med. Chem., 2025, 16, 525–544 Search PubMed.
- K. Schmidt, T. P. Prakash, A. J. Donner, G. A. Kinberger, H. J. Gaus, A. Low, M. E. Østergaard, M. Bell, E. E. Swayze and P. P. Seth, Characterizing the effect of GalNAc and phosphorothioate backbone on binding of antisense oligonucleotides to the asialoglycoprotein receptor, Nucleic Acids Res., 2017, 45, 2294–2306 CrossRef CAS PubMed.
- G. Vasquez, G. C. Freestone, W. B. Wan, A. Low, C. L. De Hoyos, J. Yu, T. P. Prakash, M. E. Ǿstergaard, X.-H. Liang, S. T. Crooke, E. E. Swayze, M. T. Migawa and P. P. Seth, Site-specific incorporation of 5′-methyl DNA enhances the therapeutic profile of gapmer ASOs, Nucleic Acids Res., 2021, 49, 1828–1839 Search PubMed.
- D. Baek, J. Villén, C. Shin, F. D. Camargo, S. P. Gygi and D. P. Bartel, The impact of microRNAs on protein output, Nature, 2008, 455, 64–71 CrossRef CAS PubMed.
- G. Hutvágner and P. D. Zamore, A microRNA in a Multiple-Turnover RNAi Enzyme Complex, Science, 2002, 297, 2056–2060 Search PubMed.
- V. C. Auyeung, I. Ulitsky, S. E. McGeary and D. P. Bartel, Beyond Secondary Structure: Primary-Sequence Determinants License Pri-miRNA Hairpins for Processing, Cell, 2013, 152, 844–858 Search PubMed.
- S. C. Kwon, T. A. Nguyen, Y.-G. Choi, M. H. Jo, S. Hohng, V. N. Kim and J.-S. Woo, Structure of Human DROSHA, Cell, 2016, 164, 81–90 Search PubMed.
- M. S. Park, R. Araya-Secchi, J. A. Brackbill, H.-D. Phan, A. C. Kehling, E. W. Abd El-Wahab, D. M. Dayeh, M. Sotomayor and K. Nakanishi, Multidomain convergence of Argonaute during RISC assembly correlates with the formation of internal water clusters, Mol. Cell, 2019, 75, 725–740 Search PubMed.
- H.-Y. Loh, B. P. Norman, K.-S. Lai, N. M. A. N. A. Rahman, N. B. M. Alitheen and M. A. Osman, The Regulatory Role of MicroRNAs in Breast Cancer, Int. J. Mol. Sci., 2019, 20, 4940 Search PubMed.
- S. W. Eichhorn, H. Guo, S. E. McGeary, R. A. Rodriguez-Mias, C. Shin, D. Baek, S.-H. Hsu, K. Ghoshal, J. Villén and D. P. Bartel, mRNA Destabilization Is the Dominant Effect of Mammalian MicroRNAs by the Time Substantial Repression Ensues, Mol. Cell, 2014, 56, 104–115 Search PubMed.
- R. S. Pillai, S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand and W. Filipowicz, Inhibition of Translational Initiation by Let-7 MicroRNA in Human Cells, Science, 2005, 309, 1573–1576 CrossRef CAS PubMed.
- C. P. Petersen, M.-E. Bordeleau, J. Pelletier and P. A. Sharp, Short RNAs Repress Translation after Initiation in Mammalian Cells, Mol. Cell, 2006, 21, 533–542 Search PubMed.
- M. R. Fabian, M. K. Cieplak, F. Frank, M. Morita, J. Green, T. Srikumar, B. Nagar, T. Yamamoto, B. Raught, T. F. Duchaine and N. Sonenberg, miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT, Nat. Struct. Mol. Biol., 2011, 18, 1211–1217 CrossRef CAS PubMed.
- L. He, J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon and S. M. Hammond, A microRNA polycistron as a potential human oncogene, Nature, 2005, 435, 828–833 CrossRef CAS PubMed.
- P. P. Medina, M. Nolde and F. J. Slack, OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma, Nature, 2010, 467, 86–90 CrossRef CAS PubMed.
- M. L. Si, S. Zhu, H. Wu, Z. Lu, F. Wu and Y. Y. Mo, miR-21-mediated tumor growth, Oncogene, 2007, 26, 2799–2803 CrossRef CAS PubMed.
- M. V. Iorio, M. Ferracin, C.-G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Ménard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini and C. M. Croce, MicroRNA Gene Expression Deregulation in Human Breast Cancer, Cancer Res., 2005, 65, 7065–7070 Search PubMed.
- C. H. Lawrie, S. Soneji, T. Marafioti, C. D. O. Cooper, S. Palazzo, J. C. Paterson, H. Cattan, T. Enver, R. Mager, J. Boultwood, J. S. Wainscoat and C. S. R. Hatton, Microrna expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma, Int. J. Cancer, 2007, 121, 1156–1161 CrossRef CAS PubMed.
- M. Jongen-Lavrencic, S. M. Sun, M. K. Dijkstra, P. J. M. Valk and B. Löwenberg, MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia, Blood, 2008, 111, 5078–5085 Search PubMed.
- X. Ma, D. J. Conklin, F. Li, Z. Dai, X. Hua, Y. Li, Z. Y. Xu-Monette, K. H. Young, W. Xiong, M. Wysoczynski, S. D. Sithu, S. Srivastava, A. Bhatnagar and Y. Li, The oncogenic microRNA miR-21 promotes regulated necrosis in mice, Nat. Commun., 2015, 6, 7151 CrossRef PubMed.
- X. Zhang, W. Chen, S. Wan, B. Qu, F. Liao, D. Cheng, Y. Zhang, Z. Ding, Y. Yang and Q. Yuan, Spatially Selective MicroRNA Imaging in Human Colorectal Cancer Tissues Using a Multivariate-Gated Signal Amplification Nanosensor, J. Am. Chem. Soc., 2025, 147, 6679–6687 Search PubMed.
- Y. Ma, N. Shen, M. S. Wicha and M. Luo, The roles of the Let-7 family of MicroRNAs in the regulation of cancer stemness, Cells, 2021, 10, 2415 CrossRef CAS PubMed.
- J. Takamizawa, H. Konishi, K. Yanagisawa, S. Tomida, H. Osada, H. Endoh, T. Harano, Y. Yatabe, M. Nagino, Y. Nimura, T. Mitsudomi and T. Takahashi, Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival, Cancer Res., 2004, 64, 3753–3756 CrossRef CAS PubMed.
- D. W. Thomson, C. P. Bracken, J. M. Szubert and G. J. Goodall, On Measuring miRNAs after Transient Transfection of Mimics or Antisense Inhibitors, PLoS One, 2013, 8, e55214 CrossRef CAS PubMed.
- T.-H. Lee, B. Matta, B. D. King, M. R. Hodges, H. L. Tillmann and K. Patel, MicroRNA-122 associates with serum apolipoprotein B but not liver fibrosis markers in CHC genotype 1 infection, J. Med. Virol., 2015, 87, 1722–1726 Search PubMed.
- S. Ottosen, T. B. Parsley, L. Yang, K. Zeh, L.-J. v. Doorn, E. v. d. Veer, A. K. Raney, M. R. Hodges and A. K. Patick, Antiviral Activity and Preclinical and Clinical Resistance Profile of Miravirsen, a Novel Anti-Hepatitis C Virus Therapeutic Targeting the Human Factor miR-122, Antimicrob. Agents Chemother., 2015, 59, 599–608 CrossRef PubMed.
- H. Hermeking, p53 Enters the MicroRNA World, Cancer Cell, 2007, 12, 414–418 Search PubMed.
- D. S. Hong, Y.-K. Kang, M. Borad, J. Sachdev, S. Ejadi, H. Y. Lim, A. J. Brenner, K. Park, J.-L. Lee, T.-Y. Kim, S. Shin, C. R. Becerra, G. Falchook, J. Stoudemire, D. Martin, K. Kelnar, H. Peltier, V. Bonato, A. G. Bader, S. Smith, S. Kim, V. O'Neill and M. S. Beg, Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours, Br. J. Cancer, 2020, 122, 1630–1637 CrossRef CAS PubMed.
- H. J. Peltier, K. Kelnar and A. G. Bader, Effects of MRX34, a liposomal miR-34 mimic, on target gene expression in human white blood cells (hWBCs): qRT-PCR results from a first-in-human trial of microRNA cancer therapy, Ann. Oncol., 2016, 27, vi531 CrossRef.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- L. Zhang, Y. Liang, G. Liang, Z. Tian, Y. Zhang, Z. Liu and X. Ji, The therapeutic prospects of N-acetylgalactosamine-siRNA conjugates, Front. Pharmacol, 2022, 13, 1090237 CrossRef CAS PubMed.
- H. Zhang, F. Ding, Z. Zhu, Q. Sun and C. Yang, Engineered ionizable lipid nanoparticles mediated efficient siRNA delivery to macrophages for anti-inflammatory treatment of acute liver injury, Int. J. Pharm., 2023, 631, 122489 CrossRef CAS PubMed.
- J. K. Nair, J. L. S. Willoughby, A. Chan, K. Charisse, M. R. Alam, Q. Wang, M. Hoekstra, P. Kandasamy, A. V. Kel’in, S. Milstein, N. Taneja, J. O'Shea, S. Shaikh, L. Zhang, R. J. van der Sluis, M. E. Jung, A. Akinc, R. Hutabarat, S. Kuchimanchi, K. Fitzgerald, T. Zimmermann, T. J. C. van Berkel, M. A. Maier, K. G. Rajeev and M. Manoharan, Multivalent N-Acetylgalactosamine-Conjugated siRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing, J. Am. Chem. Soc., 2014, 136, 16958–16961 CrossRef CAS PubMed.
- M. Kremyanskaya, R. Hoffman, L. P. Chew, A. Sharif, B. Semov, A. N. Kori, H. Sia, C. S. Grove, A. P. Ng, A. Kalro, I. Raman, G. Kasinathan, O. Oduwole, M. Gomez-Palou, H. Fok, S. Romano, C. Rambaran and A. Martinez, Initial Results from a Phase 1/2 Study Evaluating Divesiran, a Novel Galnac Conjugated siRNA, in Patients with Polycythemia Vera (SANRECO), Blood, 2024, 144, 656 CrossRef.
- J. Gilleron, W. Querbes, A. Zeigerer, A. Borodovsky, G. Marsico, U. Schubert, K. Manygoats, S. Seifert, C. Andree, M. Stöter, H. Epstein-Barash, L. Zhang, V. Koteliansky, K. Fitzgerald, E. Fava, M. Bickle, Y. Kalaidzidis, A. Akinc, M. Maier and M. Zerial, Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape, Nat. Biotechnol., 2013, 31, 638–646 CrossRef CAS PubMed.
- A. Wittrup, A. Ai, X. Liu, P. Hamar, R. Trifonova, K. Charisse, M. Manoharan, T. Kirchhausen and J. Lieberman, Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown, Nat. Biotechnol., 2015, 33, 870–876 CrossRef CAS PubMed.
- N. T. Schirle, J. Sheu-Gruttadauria and I. J. MacRae, Structural basis for microRNA targeting, Science, 2014, 346, 608–613 Search PubMed.
- M. M. Janas, M. K. Schlegel, C. E. Harbison, V. O. Yilmaz, Y. Jiang, R. Parmar, I. Zlatev, A. Castoreno, H. Xu, S. Shulga-Morskaya, K. G. Rajeev, M. Manoharan, N. D. Keirstead, M. A. Maier and V. Jadhav, Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity, Nat. Commun., 2018, 9, 723 CrossRef PubMed.
- A. K. Singh, M. Kumar, D. Choudhary, L. Aher, J. Rane and N. P. Singh, in Biotechnologies of Crop Improvement: Transgenic Approaches, Springer, 2018, vol. 2, pp. 113–127 Search PubMed.
- T. Coelho, D. Adams, A. Silva, P. Lozeron, P. N. Hawkins, T. Mant, J. Perez, J. Chiesa, S. Warrington, E. Tranter, M. Munisamy, R. Falzone, J. Harrop, J. Cehelsky, B. R. Bettencourt, M. Geissler, J. S. Butler, A. Sehgal, R. E. Meyers, Q. Chen, T. Borland, R. M. Hutabarat, V. A. Clausen, R. Alvarez, K. Fitzgerald, C. Gamba-Vitalo, S. V. Nochur, A. K. Vaishnaw, D. W. Y. Sah, J. A. Gollob and O. B. Suhr, Safety and Efficacy of RNAi Therapy for Transthyretin Amyloidosis, N. Engl. J. Med., 2013, 369, 819–829 CrossRef CAS PubMed.
- D. Adams, I. L. Tournev, M. S. Taylor, T. Coelho, V. Planté-Bordeneuve, J. L. Berk, A. González-Duarte, J. D. Gillmore, S.-C. Low, Y. Sekijima, L. Obici, C. Chen, P. Badri, S. M. Arum, J. Vest and M. Polydefkis, Efficacy and safety of vutrisiran for patients with hereditary transthyretin-mediated amyloidosis with polyneuropathy: a randomized clinical trial, Amyloid, 2023, 30, 18–26 CrossRef CAS PubMed.
- M. Balwani, E. Sardh, P. Ventura, P. A. Peiró, D. C. Rees, U. Stölzel, D. M. Bissell, H. L. Bonkovsky, J. Windyga, K. E. Anderson, C. Parker, S. M. Silver, S. B. Keel, J.-D. Wang, P. E. Stein, P. Harper, D. Vassiliou, B. Wang, J. Phillips, A. Ivanova, J. G. Langendonk, R. Kauppinen, E. Minder, Y. Horie, C. Penz, J. Chen, S. Liu, J. J. Ko, M. T. Sweetser, P. Garg, A. Vaishnaw, J. B. Kim, A. R. Simon and L. Gouya, Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria, N. Engl. J. Med., 2020, 382, 2289–2301 CrossRef CAS PubMed.
- C. Kang, Lumasiran: A Review in Primary Hyperoxaluria Type 1, Drugs, 2024, 84, 219–226 Search PubMed.
- K. K. Ray, U. Landmesser, L. A. Leiter, D. Kallend, R. Dufour, M. Karakas, T. Hall, R. P. T. Troquay, T. Turner, F. L. J. Visseren, P. Wijngaard, R. S. Wright and J. J. P. Kastelein, Inclisiran in Patients at High Cardiovascular Risk with Elevated LDL Cholesterol, N. Engl. J. Med., 2017, 376, 1430–1440 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Knocking down barriers: advances in siRNA delivery, Nat. Rev. Drug Discovery, 2009, 8, 129–138 Search PubMed.
- S. S. Ali Zaidi, F. Fatima, S. A. Ali Zaidi, D. Zhou, W. Deng and S. Liu, Engineering siRNA therapeutics: challenges and strategies, J. Nanobiotechnol., 2023, 21, 381 Search PubMed.
- N. S. Petrova, M. I. Meschaninova, A. G. Venyaminova, M. A. Zenkova, V. V. Vlassov and E. L. Chernolovskaya, Silencing activity of 2′-O-methyl modified anti-MDR1 siRNAs with mismatches in the central part of the duplexes, FEBS Lett., 2011, 585, 2352–2356 CrossRef CAS PubMed.
- Q. Li, M. Dong and P. Chen, Advances in structural-guided modifications of siRNA, Bioorg. Med. Chem., 2024, 110, 117825 CrossRef CAS PubMed.
- J. B. Bramsen, M. M. Pakula, T. B. Hansen, C. Bus, N. Langkjær, D. Odadzic, R. Smicius, S. L. Wengel, J. Chattopadhyaya, J. W. Engels, P. Herdewijn, J. Wengel and J. Kjems, A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects, Nucleic Acids Res., 2010, 38, 5761–5773 CrossRef CAS PubMed.
- M. K. Schlegel, M. M. Janas, Y. Jiang, J. D. Barry, W. Davis, S. Agarwal, D. Berman, C. R. Brown, A. Castoreno, S. LeBlanc, A. Liebow, T. Mayo, S. Milstein, T. Nguyen, S. Shulga-Morskaya, S. Hyde, S. Schofield, J. Szeto, L. B. Woods, V. O. Yilmaz, M. Manoharan, M. Egli, K. Charissé, L. Sepp-Lorenzino, P. Haslett, K. Fitzgerald, V. Jadhav and M. A. Maier, From bench to bedside: Improving the clinical safety of GalNAc–siRNA conjugates using seed-pairing destabilization, Nucleic Acids Res., 2022, 50, 6656–6670 CrossRef CAS PubMed.
- K. Fitzgerald, M. Frank-Kamenetsky, S. Shulga-Morskaya, A. Liebow, B. R. Bettencourt, J. E. Sutherland, R. M. Hutabarat, V. A. Clausen, V. Karsten, J. Cehelsky, S. V. Nochur, V. Kotelianski, J. Horton, T. Mant, J. Chiesa, J. Ritter, M. Munisamy, A. K. Vaishnaw, J. A. Gollob and A. Simon, Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial, Lancet, 2014, 383, 60–68 Search PubMed.
- O. Alkhamis and Y. Xiao, Systematic Study of in Vitro Selection Stringency Reveals How to Enrich High-Affinity Aptamers, J. Am. Chem. Soc., 2023, 145, 194–206 Search PubMed.
- Y. Song, J. Song, X. Wei, M. Huang, M. Sun, L. Zhu, B. Lin, H. Shen, Z. Zhu and C. Yang, Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein, Anal. Chem., 2020, 92, 9895–9900 Search PubMed.
- S. Wu, Y. Shang, Y. Yan, A. Zhou, T. Bing, Z. Zhao and W. Tan, Aptamer-based enforced phosphatase-recruiting chimeras inhibit receptor tyrosine kinase signal transduction, J. Am. Chem. Soc., 2024, 146, 22445–22454 CrossRef CAS PubMed.
- Y. Sima, L. Ai, L. Wang, P. Zhang, Q. Zhang, S. Wu, S. Xie, Z. Zhao and W. Tan, A DNA Molecular Logic Circuit for Precise Tumor Identification, Nano Lett., 2024, 24, 12070–12079 Search PubMed.
- S. Wu, X. Zhou, L. Ai, Y. Shang, Y. Yan, Z. Zhao and W. Tan, A Cyclized Bivalent Aptamer-Based Protein Degrader Targeting Receptor Tyrosine Kinases Overcomes Resistance to Inhibitors in Lung Cancer, ACS Nano, 2025, 19, 28171–28185 CrossRef CAS PubMed.
- L. Ai, Q. Zuo, Y. Li, S. Wu, Y. Sima, Y. Zhang, S. Xie, Z. Zhao and W. Tan, Programming Affinity for Precise Tumor Recognition with Allosteric Nanosensing-Circles, ACS Nano, 2023, 17, 13430–13440 CrossRef CAS PubMed.
- F. Zhou, P. Wang, J. Chen, Z. Zhu, Y. Li, S. Wang, S. Wu, Y. Sima, T. Fu and W. Tan, A photochemically covalent lock stabilizes aptamer conformation and strengthens its performance for biomedicine, Nucleic Acids Res., 2022, 50, 9039–9050 CrossRef CAS PubMed.
- J. O. McNamara, E. R. Andrechek, Y. Wang, K. D. Viles, R. E. Rempel, E. Gilboa, B. A. Sullenger and P. H. Giangrande, Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras, Nat. Biotechnol., 2006, 24, 1005–1015 Search PubMed.
- C.-H. B. Chen, K. R. Dellamaggiore, C. P. Ouellette, C. D. Sedano, M. Lizadjohry, G. A. Chernis, M. Gonzales, F. E. Baltasar, A. L. Fan, R. Myerowitz and E. F. Neufeld, Aptamer-based endocytosis of a lysosomal enzyme, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15908–15913 CrossRef CAS PubMed.
- G. Zhu, G. Niu and X. Chen, Aptamer–drug conjugates, Bioconjug. Chem., 2015, 26, 2186–2197 CrossRef CAS PubMed.
- V. Bagalkot, O. C. Farokhzad, R. Langer and S. Jon, An Aptamer–Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform, Angew. Chem., Int. Ed., 2006, 45, 8149–8152 CrossRef CAS PubMed.
- F. Li, J. Lu, J. Liu, C. Liang, M. Wang, L. Wang, D. Li, H. Yao, Q. Zhang, J. Wen, Z.-K. Zhang, J. Li, Q. Lv, X. He, B. Guo, D. Guan, Y. Yu, L. Dang, X. Wu, Y. Li, G. Chen, F. Jiang, S. Sun, B.-T. Zhang, A. Lu and G. Zhang, A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer, Nat. Commun., 2017, 8, 1390 CrossRef PubMed.
- S. Kruspe and U. Hahn, An Aptamer Intrinsically Comprising 5-Fluoro-2′-deoxyuridine for Targeted Chemotherapy, Angew. Chem., Int. Ed., 2014, 53, 10541–10544 Search PubMed.
- B.-T. Huang, W.-Y. Lai, C.-L. Yeh, Y.-T. Tseng, K. Peck, P.-C. Yang and E. P.-Y. Lin, AptBCis1, An Aptamer–Cisplatin Conjugate, Is Effective in Lung Cancer Leptomeningeal Carcinomatosis, ACS Nano, 2024, 18, 27905–27916 CrossRef CAS PubMed.
- C. Luo, X. Hu, R. Peng, H. Huang, Q. Liu and W. Tan, Biomimetic Carriers Based on Giant Membrane Vesicles for Targeted Drug Delivery and Photodynamic/Photothermal Synergistic Therapy, ACS Appl. Mater. Interfaces, 2019, 11, 43811–43819 CrossRef CAS.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Quadruplex DNA: sequence, topology and structure, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed.
- Y. Otake, S. Soundararajan, T. K. Sengupta, E. A. Kio, J. C. Smith, M. Pineda-Roman, R. K. Stuart, E. K. Spicer and D. J. Fernandes, Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA, Blood, 2007, 109, 3069–3075 CrossRef CAS PubMed.
- C.-h. B. Chen, G. A. Chernis, V. Q. Hoang and R. Landgraf, Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3, Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 9226–9231 CrossRef CAS PubMed.
- F. Pastor, M. M. Soldevilla, H. Villanueva, D. Kolonias, S. Inoges, A. L. de Cerio, R. Kandzia, V. Klimyuk, Y. Gleba, E. Gilboa and M. Bendandi, CD28 Aptamers as Powerful Immune Response Modulators, Mol. Ther. Nucleic Acids, 2013, 2, e98 CrossRef PubMed.
- C. M. Dollins, S. Nair, D. Boczkowski, J. Lee, J. M. Layzer, E. Gilboa and B. A. Sullenger, Assembling OX40 Aptamers on a Molecular Scaffold to Create a Receptor-Activating Aptamer, Chem. Biol., 2008, 15, 675–682 CrossRef CAS PubMed.
- M. M. Soldevilla, H. Villanueva, M. Bendandi, S. Inoges, A. López-Díaz de Cerio and F. Pastor, 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia, Biomaterials, 2015, 67, 274–285 CrossRef CAS PubMed.
- V. Ramaswamy, A. Monsalve, L. Sautina, M. S. Segal, J. Dobson and J. B. Allen, DNA Aptamer Assembly as a Vascular Endothelial Growth Factor Receptor Agonist, Nucleic Acid Ther., 2015, 25, 227–234 Search PubMed.
- N.-O. Yunn, A. Koh, S. Han, J. H. Lim, S. Park, J. Lee, E. Kim, S. K. Jang, P.-O. Berggren and S. H. Ryu, Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation, Nucleic Acids Res., 2015, 43, 7688–7701 Search PubMed.
- E. W. M. Ng, D. T. Shima, P. Calias, E. T. Cunningham, D. R. Guyer and A. P. Adamis, Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease, Nat. Rev. Drug Discovery, 2006, 5, 123–132 CrossRef CAS PubMed.
- G. J. Jaffe, K. Westby, K. G. Csaky, J. Monés, J. A. Pearlman, S. S. Patel, B. C. Joondeph, J. Randolph, H. Masonson and K. A. Rezaei, C5 Inhibitor Avacincaptad Pegol for Geographic Atrophy Due to Age-Related Macular Degeneration: A Randomized Pivotal Phase 2/3 Trial, Ophthalmology, 2021, 128, 576–586 CrossRef PubMed.
- C. Kratschmer and M. Levy, Effect of Chemical Modifications on Aptamer Stability in Serum, Nucleic Acid Ther., 2017, 27, 335–344 CrossRef CAS PubMed.
- M. Egli and M. Manoharan, Re-Engineering RNA Molecules into Therapeutic Agents, Acc. Chem. Res., 2019, 52, 1036–1047 CrossRef CAS PubMed.
- Q. Yang, Y. Peng, Z. Deng, D. Zhang, C.-Y. Long, G.-R. Zhang, J. Li, X.-Q. Wang and W. Tan, Regulating the properties of XQ-2d for targeted delivery of therapeutic agents to pancreatic cancers, Natl. Sci. Rev., 2023, 10, nwad113 CrossRef CAS PubMed.
- I. C. Elle, K. K. Karlsen, M. G. Terp, N. Larsen, R. Nielsen, N. Derbyshire, S. Mandrup, H. J. Ditzel and J. Wengel, Selection of LNA-containing DNA aptamers against recombinant human CD73, Mol. BioSyst., 2015, 11, 1260–1270 Search PubMed.
- A. Prodeus, A. Abdul-Wahid, N. W. Fischer, E. H. B. Huang, M. Cydzik and J. Gariépy, Targeting the PD-1/PD-L1 Immune Evasion Axis with DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers, Mol. Ther. Nucleic Acids, 2015, 4, e237 Search PubMed.
- C. Maasch, K. Buchner, D. Eulberg, S. Vonhoff and S. Klussmann, Physicochemical Stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for Therapeutic Applications, Nucleic Acids Symp. Ser., 2008, 52, 61–62 CrossRef CAS PubMed.
- C. Ji, J. Wei, L. Zhang, X. Hou, J. Tan, Q. Yuan and W. Tan, Aptamer–Protein Interactions: From Regulation to Biomolecular Detection, Chem. Rev., 2023, 123, 12471–12506 CrossRef CAS.
- C. Riccardi, A. Meyer, J.-J. Vasseur, I. Russo Krauss, L. Paduano, F. Morvan and D. Montesarchio, Fine-tuning the properties of the thrombin binding aptamer through cyclization: Effect of the 5′-3′ connecting linker on the aptamer stability and anticoagulant activity, Bioorg. Chem., 2020, 94, 103379 CrossRef CAS PubMed.
- L. Zhao, X. Qi, X. Yan, Y. Huang, X. Liang, L. Zhang, S. Wang and W. Tan, Engineering Aptamer with Enhanced Affinity by Triple Helix-Based Terminal Fixation, J. Am. Chem. Soc., 2019, 141, 17493–17497 CrossRef CAS PubMed.
- J. A. Wolff, R. W. Malone, P. Williams, W. Chong, G. Acsadi, A. Jani and P. L. Felgner, Direct Gene Transfer into Mouse Muscle in Vivo, Science, 1990, 247, 1465–1468 CrossRef CAS PubMed.
- K. Karikó, M. Buckstein, H. Ni and D. Weissman, Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA, Immunity, 2005, 23, 165–175 Search PubMed.
- K. Karikó, H. Muramatsu, F. A. Welsh, J. Ludwig, H. Kato, S. Akira and D. Weissman, Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability, Mol. Ther., 2008, 16, 1833–1840 CrossRef PubMed.
- R. Verbeke, I. Lentacker, S. C. De Smedt and H. Dewitte, The dawn of mRNA vaccines: The COVID-19 case, J. Controlled Release, 2021, 333, 511–520 CrossRef CAS PubMed.
- Y. N. Lamb, BNT162b2 mRNA COVID-19 Vaccine: First Approval, Drugs, 2021, 81, 495–501 CrossRef CAS PubMed.
- A. Sfera, K. Thomas, D. Sfera, J. Anton, C. Andronescu, N. Jafri, S. Susanna and Z. Kozlakidis, Do messenger RNA vaccines induce pathological syncytial, Int. J. Pathol. Clin. Res, 2022, 8, 137 Search PubMed.
- J. C. Muodiaju, C. S. Madu and J. Muodiaju, Messenger Ribonucleic Acid (mRNA)-Based Universal Vaccines: Engineering Broad-Spectrum Immunity Against Future Pandemics, Cureus, 2025, 17, e84821 Search PubMed.
- R. N. Germain, Immunology: the ins and outs of antigen processing and presentations, Nature, 1986, 322, 687–688 CrossRef CAS PubMed.
- A. Townsend, T. Elliott, V. Cerundolo, L. Foster, B. Barber and A. Tse, Assembly of MHC class I molecules analyzed in vitro, Cell, 1990, 62, 285–295 CrossRef CAS PubMed.
- U. Sahin, A. Muik, E. Derhovanessian, I. Vogler, L. M. Kranz, M. Vormehr, A. Baum, K. Pascal, J. Quandt, D. Maurus, S. Brachtendorf, V. Lörks, J. Sikorski, R. Hilker, D. Becker, A.-K. Eller, J. Grützner, C. Boesler, C. Rosenbaum, M.-C. Kühnle, U. Luxemburger, A. Kemmer-Brück, D. Langer, M. Bexon, S. Bolte, K. Karikó, T. Palanche, B. Fischer, A. Schultz, P.-Y. Shi, C. Fontes-Garfias, J. L. Perez, K. A. Swanson, J. Loschko, I. L. Scully, M. Cutler, W. Kalina, C. A. Kyratsous, D. Cooper, P. R. Dormitzer, K. U. Jansen and Ö. Türeci, COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses, Nature, 2020, 586, 594–599 Search PubMed.
- L. A. Jackson, E. J. Anderson, N. G. Rouphael, P. C. Roberts, M. Makhene, R. N. Coler, M. P. McCullough, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. McDermott, B. Flach, N. A. Doria-Rose, K. S. Corbett, K. M. Morabito, S. O'Dell, S. D. Schmidt, P. A. Swanson, M. Padilla, J. R. Mascola, K. M. Neuzil, H. Bennett, W. Sun, E. Peters, M. Makowski, J. Albert, K. Cross, W. Buchanan, R. Pikaart-Tautges, J. E. Ledgerwood, B. S. Graham and J. H. Beigel, An mRNA Vaccine against SARS-CoV-2—Preliminary Report, N. Engl. J. Med., 2020, 383, 1920–1931 CrossRef CAS PubMed.
- G. L. Chen, R. Mithani, A. Kapoor, S. Lu, L. E. Asmar, C. A. Panozzo, C. A. Shaw, S. K. Stoszek and A. August, Safety and Immunogenicity of mRNA-1345, an mRNA-Based RSV Vaccine in Younger and Older Adult Cohorts: Results from a Phase 1, Randomized Clinical Trial, Open Forum Infect. Dis., 2022, 9, ofac492.312 Search PubMed.
- A. O. Aliprantis, C. A. Shaw, P. Griffin, N. Farinola, R. A. Railkar, X. Cao, W. Liu, J. R. Sachs, C. J. Swenson, H. Lee, K. S. Cox, D. S. Spellman, C. J. Winstead, I. Smolenov, E. Lai, T. Zaks, A. S. Espeseth and L. Panther, A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults, Hum. Vaccines Immunother., 2021, 17, 1248–1261 Search PubMed.
- M. Soens, J. Ananworanich, B. Hicks, K. J. Lucas, J. Cardona, L. Sher, G. Livermore, K. Schaefers, C. Henry, A. Choi, A. Avanesov, R. Chen, E. Du, A. Pucci, R. Das, J. Miller and R. Nachbagauer, A phase 3 randomized safety and immunogenicity trial of mRNA-1010 seasonal influenza vaccine in adults, Vaccine, 2025, 50, 126847 CrossRef CAS PubMed.
- K. Bahl, J. J. Senn, O. Yuzhakov, A. Bulychev, L. A. Brito, K. J. Hassett, M. E. Laska, M. Smith, Ö. Almarsson, J. Thompson, A. Ribeiro, M. Watson, T. Zaks and G. Ciaramella, Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses, Mol. Ther., 2017, 25, 1316–1327 CrossRef CAS PubMed.
- J. S. Weber, J. J. Luke, M. S. Carlino, M. A. Khattak, R. S. Meehan, M. Brown, J. Zhang, C. Krepler, J. P. Duic and G. V. Long, INTerpath-001: Pembrolizumab with V940 (mRNA-4157) versus pembrolizumab with placebo for adjuvant treatment of high-risk stage II–IV melanoma, J. Clin. Oncol., 2024, 42, TPS9616 Search PubMed.
- P. M. Forde, J. E. Chaft, K. N. Smith, V. Anagnostou, T. R. Cottrell, M. D. Hellmann, M. Zahurak, S. C. Yang, D. R. Jones, S. Broderick, R. J. Battafarano, M. J. Velez, N. Rekhtman, Z. Olah, J. Naidoo, K. A. Marrone, F. Verde, H. Guo, J. Zhang, J. X. Caushi, H. Y. Chan, J.-W. Sidhom, R. B. Scharpf, J. White, E. Gabrielson, H. Wang, G. L. Rosner, V. Rusch, J. D. Wolchok, T. Merghoub, J. M. Taube, V. E. Velculescu, S. L. Topalian, J. R. Brahmer and D. M. Pardoll, Neoadjuvant PD-1 Blockade in Resectable Lung Cancer, N. Engl. J. Med., 2018, 378, 1976–1986 Search PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing, Nat. Nanotechnol., 2020, 15, 313–320 Search PubMed.
- A. Thess, S. Grund, B. L. Mui, M. J. Hope, P. Baumhof, M. Fotin-Mleczek and T. Schlake, Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals, Mol. Ther., 2015, 23, 1456–1464 Search PubMed.
- T. Vukovich, K. Auberger, J. Weil, H. Engelmann, P. Knöbl and H. B. Hadorn, Replacement therapy for a homozygous protein C deficiency-state using a concentrate of human protein C and S, Br. J. Haematol., 1988, 70, 435–440 Search PubMed.
- T. Vavilis, E. Stamoula, A. Ainatzoglou, A. Sachinidis, M. Lamprinou, I. Dardalas and I. S. Vizirianakis, mRNA in the Context of Protein Replacement Therapy, Pharmaceutics, 2023, 15 CrossRef CAS PubMed.
- S. A. Dilliard and D. J. Siegwart, Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- D. An, J. L. Schneller, A. Frassetto, S. Liang, X. Zhu, J. S. Park, M. Theisen, S. J. Hong, J. Zhou, R. Rajendran, B. Levy, R. Howell, G. Besin, V. Presnyak, S. Sabnis, K. E. Murphy-Benenato, E. S. Kumarasinghe, T. Salerno, C. Mihai, C. M. Lukacs, R. J. Chandler, L. T. Guey, C. P. Venditti and P. G. V. Martini, Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia, Cell Rep., 2017, 21, 3548–3558 CrossRef CAS PubMed.
- D. Koeberl, A. Schulze, N. Sondheimer, G. S. Lipshutz, T. Geberhiwot, L. Li, R. Saini, J. Luo, V. Sikirica, L. Jin, M. Liang, M. Leuchars and S. Grunewald, Interim analyses of a first-in-human phase 1/2 mRNA trial for propionic acidaemia, Nature, 2024, 628, 872–877 Search PubMed.
- K. Yamazaki, K. Kubara, S. Ishii, K. Kondo, Y. Suzuki, T. Miyazaki, K. Mitsuhashi, M. Ito and K. Tsukahara, Lipid nanoparticle-targeted mRNA formulation as a treatment for ornithine-transcarbamylase deficiency model mice, Mol. Ther. Nucleic Acids, 2023, 33, 210–226 Search PubMed.
- M. Laurent, M. Geoffroy, G. Pavani and S. Guiraud, CRISPR-Based Gene Therapies: From Preclinical to Clinical Treatments, Cells, 2024, 13 Search PubMed.
- J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana, M. L. Maitland, J. Seitzer, D. O'Connell, K. R. Walsh, K. Wood, J. Phillips, Y. Xu, A. Amaral, A. P. Boyd, J. E. Cehelsky, M. D. McKee, A. Schiermeier, O. Harari, A. Murphy, C. A. Kyratsous, B. Zambrowicz, R. Soltys, D. E. Gutstein, J. Leonard, L. Sepp-Lorenzino and D. Lebwohl, CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis, N. Engl. J. Med., 2021, 385, 493–502 Search PubMed.
- V. Anttila, A. Saraste, J. Knuuti, M. Hedman, P. Jaakkola, K. L. Laugwitz, M. Krane, A. Jeppsson, S. Sillanmäki, J. Rosenmeier, P. Zingmark, A. Rudvik, P. Garkaviy, C. Watson, M. N. Pangalos, K. R. Chien, R. Fritsche-Danielson, A. Collén and L. M. Gan, Direct intramyocardial injection of VEGF mRNA in patients undergoing coronary artery bypass grafting, Mol. Ther. Nucleic Acids, 2023, 31, 866–874 CAS.
- M. Kwak and A. Herrmann, Nucleic acid amphiphiles: synthesis and self-assembled nanostructures, Chem. Soc. Rev., 2011, 40, 5745–5755 Search PubMed.
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99, 237–247 Search PubMed.
- D.-X. Wang, J. Wang, Y.-X. Wang, Y.-C. Du, Y. Huang, A.-N. Tang, Y.-X. Cui and D.-M. Kong, DNA nanostructure-based nucleic acid probes: construction and biological applications, Chem. Sci., 2021, 12, 7602–7622 RSC.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, Spherical nucleic acids, J. Am. Chem. Soc., 2012, 134, 1376–1391 CrossRef CAS PubMed.
- S. P. Narayan, C. H. J. Choi, L. Hao, C. M. Calabrese, E. Auyeung, C. Zhang, O. J. Goor and C. A. Mirkin, The sequence-specific cellular uptake of spherical nucleic acid nanoparticle conjugates, Small, 2015, 11, 4173–4182 Search PubMed.
- H. Li, B. Zhang, X. Lu, X. Tan, F. Jia, Y. Xiao, Z. Cheng, Y. Li, D. O. Silva and H. S. Schrekker, Molecular spherical nucleic acids, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 4340–4344 Search PubMed.
- M.-E. Kyriazi, A. H. El-Sagheer, I. L. Medintz, T. Brown and A. G. Kanaras, An investigation into the resistance of spherical nucleic acids against DNA enzymatic degradation, Bioconjugate Chem., 2022, 33, 219–225 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Cellular uptake of nanoparticles: journey inside the cell, Chem. Soc. Rev., 2017, 46, 4218–4244 Search PubMed.
- Y. Song, W. Song, X. Lan, W. Cai and D. Jiang, Spherical nucleic acids: Organized nucleotide aggregates as versatile nanomedicine, Aggregate, 2022, 3, e120 Search PubMed.
- J. Li, X. Mao, T. Zhao, W. Fang, Y. Jin, M. Liu, C. Fan and Y. Tian, Tetrahedral DNA Framework-Based Spherical Nucleic Acids for Efficient siRNA Delivery, Angew. Chem., Int. Ed., 2025, 137, e202416988 CrossRef.
- G. Wang, S. Han and Y. Lu, From structure to application: the evolutionary trajectory of spherical nucleic acids, Small, 2024, 20, 2310026 CrossRef CAS PubMed.
- H. Kim, H. Choi, Y. Heo, C. Kim, M. Kim and K. T. Kim, Biosensors based on bivalent and multivalent recognition by nucleic acid scaffolds, Appl. Sci., 2022, 12, 1717 Search PubMed.
- X. Wang, T. Yang, Z. Yu, T. Liu, R. Jin, L. Weng, Y. Bai, J. J. Gooding, Y. Zhang and X. Chen, Intelligent Gold Nanoparticles with Oncogenic MicroRNA-Dependent Activities to Manipulate Tumorigenic Environments for Synergistic Tumor Therapy, Adv. Mater., 2022, 34, 2110219 CrossRef CAS PubMed.
- W. L. Daniel, U. Lorch, S. Mix and A. S. Bexon, A first-in-human phase 1 study of cavrotolimod, a TLR9 agonist spherical nucleic acid, in healthy participants: evidence of immune activation, Front. Immunol., 2022, 13, 1073777 Search PubMed.
- T. R. Holmes and A. S. Paller, Correction: Holmes, TR; Paller, AS Gene Regulation Using Spherical Nucleic Acids to Treat Skin Disorders, Pharmaceuticals, 2025, 18, 1091 Search PubMed.
- S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. Nijenhuis, B. Saccà and F. C. Simmel, DNA origami, Nat. Rev. Methods Primers, 2021, 1, 13 Search PubMed.
- P. W. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 Search PubMed.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, DNA origami with complex curvatures in three-dimensional space, Science, 2011, 332, 342–346 Search PubMed.
- S. M. Douglas, H. Dietz, T. Liedl, B. Högberg, F. Graf and W. M. Shih, Self-assembly of DNA into nanoscale three-dimensional shapes, Nature, 2009, 459, 414–418 Search PubMed.
- P. Pitikultham, Z. Wang, Y. Wang, Y. Shang, Q. Jiang and B. Ding, Stimuli-responsive DNA origami nanodevices and their biological applications, ChemMedChem, 2022, 17, e202100635 Search PubMed.
- L. Li, S. Nie, T. Du, J. Zhao and X. Chen, DNA origami technology for biomedical applications: challenges and opportunities, MedComm – Biomater. Appl., 2023, 2, e37 Search PubMed.
- A. E. Marras, L. Zhou, H.-J. Su and C. E. Castro, Programmable motion of DNA origami mechanisms, Proc. Natl. Acad. Sci. U.S.A, 2015, 112, 713–718 Search PubMed.
- G. Chatterjee, N. Dalchau, R. A. Muscat, A. Phillips and G. Seelig, A spatially localized architecture for fast and modular DNA computing, Nat. Nanotechnol., 2017, 12, 920–927 Search PubMed.
- Y. Huang, W. Huang, L. Chan, B. Zhou and T. Chen, A multifunctional DNA origami as carrier of metal complexes to achieve enhanced tumoral delivery and nullified systemic toxicity, Biomaterials, 2016, 103, 183–196 Search PubMed.
- G. Li, C. Chen, Y. Li, B. Wang, J. Wen, M. Guo, M. Chen, X.-B. Zhang and G. Ke, DNA-origami-based precise molecule assembly and their biological applications, Nano Lett., 2024, 24, 11335–11348 Search PubMed.
- Z. Wang, L. Song, Q. Liu, R. Tian, Y. Shang, F. Liu, S. Liu, S. Zhao, Z. Han and J. Sun, A tubular DNA nanodevice as a siRNA/chemo-drug co-delivery vehicle for combined cancer therapy, Angew. Chem., Int. Ed., 2021, 133, 2626–2630 Search PubMed.
- J. Zhang, Y. Xu, M. Chen, Y. Huang, T. Song, C. Yang, Y. Yang and Y. Song, Elucidating the effect of nanoscale receptor-binding domain organization on SARS-CoV-2 infection and immunity activation with DNA origami, J. Am. Chem. Soc., 2022, 144, 21295–21303 Search PubMed.
- X. Hu, H. Chi, X. Fu, J. Chen, L. Dong, S. Jiang, Y. Li, J. Chen, M. Cheng and Q. Min, Tunable multivalent aptamer-based DNA nanostructures to regulate multiheteroreceptor-mediated tumor recognition, J. Am. Chem. Soc., 2024, 146, 2514–2523 Search PubMed.
- T. Xu, S. Yu, Y. Sun, S. Wu, D. Gao, M. Wang, Z. Wang, Y. Tian, Q. Min and J. J. Zhu, DNA Origami Frameworks Enabled Self-Protective siRNA Delivery for Dual Enhancement of Chemo-Photothermal Combination Therapy, Small, 2021, 17, 2101780 CrossRef CAS PubMed.
- X. Jiang, N. Wang, C. Liu, Y. Zhuo, L. Liang, Y. Gan and M. Yu, delivery of nucleic acid therapeutics: challenges, strategies, and opportunities, Drug Discov. Today, 2023, 28, 103507 Search PubMed.
- R. Zhang, R. El-Mayta, T. J. Murdoch, C. C. Warzecha, M. M. Billingsley, S. J. Shepherd, N. Gong, L. Wang, J. M. Wilson, D. Lee and M. J. Mitchell, Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver, Biomater. Sci., 2021, 9, 1449–1463 RSC.
- S. M. Kim, P. H. Faix and J. E. Schnitzer, Overcoming key biological barriers to cancer drug delivery and efficacy, J. Controlled Release, 2017, 267, 15–30 CrossRef CAS PubMed.
- Y. Ma, S. Li, X. Lin and Y. Chen, Bioinspired Spatiotemporal Management toward RNA Therapies, ACS Nano, 2023, 17, 24539–24563 Search PubMed.
- S. W. Beh CW, Y. Wang, Y. Zhang, Z. Y. Ong, P. L. Ee and Y. Y. Yang, Efficient delivery of Bcl-2-targeted siRNA using cationic polymer nanoparticles: downregulating mRNA expression level and sensitizing cancer cells to anticancer drug, Biomacromolecules, 2009, 10, 41–48 CrossRef PubMed.
- T. Tsuboi, K. Hattori, T. Ishimoto, K. Imai, T. Doke, J. Hagita, J. Ariyoshi, K. Furuhashi, N. Kato, Y. Ito, Y. Kamiya, H. Asanuma and S. Maruyama, In vivo efficacy and safety of systemically administered serinol nucleic acid-modified antisensMolecular Therapye oligonucleotides in mouse kidney, Mol. Ther. Nucleic Acids, 2025, 36, 102387 CrossRef CAS PubMed.
- A. Gardner and B. Ruffell, Dendritic Cells and Cancer Immunity, Trends Immunol., 2016, 37, 855–865 CrossRef CAS PubMed.
- A. S. Boisgard, M. Lamrayah, M. Dzikowski, D. Salmon, P. Kirilov, C. Primard, F. Pirot, B. Fromy and B. Verrier, Innovative drug vehicle for local treatment of inflammatory skin diseases: ex vivo and in vivo screening of five topical formulations containing poly(lactic acid) (PLA) nanoparticles, Eur. J. Pharm. Biopharm., 2017, 116, 51–60 CrossRef CAS PubMed.
- M. Jeong, Y. Lee, J. Park, H. Jung and H. Lee, Lipid nanoparticles (LNPs) for in vivo RNA delivery and their breakthrough technology for future applications, Adv. Drug Delivery Rev., 2023, 200, 114990 Search PubMed.
- M. Toporkiewicz, J. Meissner, L. Matusewicz, A. Czogalla and A. F. Sikorski, Toward a magic or imaginary bullet? Ligands for drug targeting to cancer cells: principles, hopes, and challenges, Int. J. Nanomed., 2015, 10, 1399–1414 Search PubMed.
- M. A. Rodger and L. King, Drawing up and administering intramuscular injections: a review of the literature, J. Adv. Nurs., 2000, 31, 574–582 CrossRef CAS PubMed.
- J. Alter, F. Lou, A. Rabinowitz, H. Yin, J. Rosenfeld, S. D. Wilton, T. A. Partridge and Q. L. Lu, Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology, Nat. Med., 2006, 12, 175–177 Search PubMed.
- L. Schoenmaker, D. Witzigmann, J. A. Kulkarni, R. Verbeke, G. Kersten, W. Jiskoot and D. J. A. Crommelin, mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability, Int. J. Pharm., 2021, 601 Search PubMed.
- P. R. Cullis and M. J. Hope, Lipid Nanoparticle Systems for Enabling Gene Therapies, Mol. Ther. Nucleic Acids, 2017, 25, 1467–1475 CAS.
- J. L. Madrid, L. V. Fatela, R. D. Lobato and A. Gozalo, Intrathecal therapy: rationale, technique, clinical results, Acta Anaesthesiol Scand, 1987, 31, 60–67 Search PubMed.
- T. Miller, M. Cudkowicz, P. J. Shaw, P. M. Andersen, N. Atassi, R. C. Bucelli, A. Genge, J. Glass, S. Ladha, A. L. Ludolph, N. J. Maragakis, C. J. McDermott, A. Pestronk, J. Ravits, F. Salachas, R. Trudell, P. Van Damme, L. Zinman, C. F. Bennett, R. Lane, A. Sandrock, H. Runz, D. Graham, H. Houshyar, A. McCampbell, I. Nestorov, I. Chang, M. McNeill, L. Fanning, S. Fradette and T. A. Ferguson, Phase 1–2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS, N. Engl. J. Med., 2020, 383, 109–119 CrossRef CAS PubMed.
- A. Mehta, A. Sonabend and J. Bruce, Convection-enhanced delivery, Neurotherapeutics, 2017, 14, 358–371 CrossRef CAS PubMed.
- M. Yoshimura, M. Sakamoto, M. Ikemoto, Y. Mochizuki, K. Yuasa, Y. Miyagoe-Suzuki and S. Takeda, AAV vector-mediated microdystrophin expression in a relatively small percentage of mdx myofibers improved the mdx phenotype, Mol. Ther. Nucleic Acids, 2004, 10, 821–828 CAS.
- J. R. Melamed, S. S. Yerneni, M. L. Arral, S. T. LoPresti, N. Chaudhary, A. Sehrawat, H. Muramatsu, M.-G. Alameh, N. Pardi, D. Weissman, G. K. Gittes and K. A. Whitehead, Ionizable lipid nanoparticles deliver mRNA to pancreatic β cells via macrophage-mediated gene transfer, Sci. Adv., 2023, 9, eade1444 Search PubMed.
- R. H. Andtbacka, H. L. Kaufman, F. Collichio, T. Amatruda, N. Senzer, J. Chesney, K. A. Delman, L. E. Spitler, I. Puzanov, S. S. Agarwala, M. Milhem, L. Cranmer, B. Curti, K. Lewis, M. Ross, T. Guthrie, G. P. Linette, G. A. Daniels, K. Harrington, M. R. Middleton, W. H. Miller Jr, J. S. Zager, Y. Ye, B. Yao, A. Li, S. Doleman, A. VanderWalde, J. Gansert and R. S. Coffin, Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma, J. Clin. Oncol., 2015, 33, 2780–2788 Search PubMed.
- M. D. de Smet, C. J. Meenken and G. J. van den Horn, Fomivirsen – a phosphorothioate oligonucleotide for the treatment of CMV retinitis, Ocul. Immunol. Inflammation, 1999, 7, 189–198 CrossRef CAS PubMed.
- J. Bennicelli, J. F. Wright, A. Komaromy, J. B. Jacobs, B. Hauck, O. Zelenaia, F. Mingozzi, D. Hui, D. Chung, T. S. Rex, Z. Wei, G. Qu, S. Zhou, C. Zeiss, V. R. Arruda, G. M. Acland, L. F. Dell'Osso, K. A. High, A. M. Maguire and J. Bennett, Reversal of blindness in animal models of leber congenital amaurosis using optimized AAV2-mediated gene transfer, Mol. Ther. Nucleic Acids, 2008, 16, 458–465 CAS.
- E. Grudzien, M. Kalek, J. Jemielity, E. Darzynkiewicz and R. E. Rhoads, Differential inhibition of mRNA degradation pathways by novel cap analogs, J. Biol. Chem., 2006, 281, 1857–1867 Search PubMed.
- H. Jiang, J. Zuo, B. Li, R. Chen, K. Luo, X. Xiang, S. Lu, C. Huang, L. Liu, J. Tang and F. Gao, Drug-induced oxidative stress in cancer treatments: Angel or devil?, Redox Biol., 2023, 63, 102754 Search PubMed.
- M. J. Kamali, M. Salehi, S. Fatemi, F. Moradi, A. Khoshghiafeh and M. Ahmadifard, Locked nucleic acid (LNA): a modern approach to cancer diagnosis and treatment, Exp. Cell Res., 2023, 423, 113442 Search PubMed.
- X. Li, S. Zhou, H. S. Tae, S. Wang, T. Li, W. Cai, T. Jiang, D. J. Adams and R. Yu, N-Terminal Capping of the alphaO-Conotoxin Analogue GeX-2 Improves the Serum Stability and Selectivity toward the Human alpha9alpha10 Nicotinic Acetylcholine Receptor, J. Med. Chem., 2024, 67, 18400–18411 CrossRef CAS PubMed.
- C. Hald Albertsen, J. A. Kulkarni, D. Witzigmann, M. Lind, K. Petersson and J. B. Simonsen, The role of lipid components in lipid nanoparticles for vaccines and gene therapy, Adv. Drug Delivery Rev., 2022, 188, 114416 CrossRef CAS PubMed.
- S. Shaikh, P. S. Chary and N. K. Mehra, Supramolecular polymers: a perspective on the stability, biocompatibility and regulatory aspects, Int. J. Pharm., 2025, 671, 125277 CrossRef CAS PubMed.
- D. E. Jensen, R. C. Kelly and P. H. von Hippel, DNA “melting” proteins. II. Effects of bacteriophage T4 gene 32-protein binding on the conformation and stability of nucleic acid structures, J. Biol. Chem., 1976, 251, 7215–7228 CrossRef CAS PubMed.
- Y. Zhang, J. Liu, C. Wang, J. Liu and W. Lu, Toll-Like Receptors Gene Polymorphisms in Autoimmune Disease, Front. Immunol., 2021, 12, 672346 CrossRef CAS PubMed.
- Y. M. Loo and M. Gale Jr, Immune signaling by RIG-I-like receptors, Immunity, 2011, 34, 680–692 CrossRef CAS PubMed.
- T. Balasubramaniyam, T. Ishizuka and Y. Xu, Stability and properties of Z-DNA containing artificial nucleobase 2'-O-methyl-8-methyl guanosine, Bioorg. Med. Chem., 2019, 27, 364–369 CrossRef CAS PubMed.
- E. Mao and D. W. C. MacMillan, Late-Stage C(sp(3))–H Methylation of Drug Molecules, J. Am. Chem. Soc., 2023, 145, 2787–2793 CrossRef CAS PubMed.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, Lipid Nanoparticles horizontal line From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS PubMed.
- H. X. Deng, H. Zhai, Y. Shi, G. Liu, J. Lowry, B. Liu, E. B. Ryan, J. Yan, Y. Yang, N. Zhang, Z. Yang, E. Liu, Y. C. Ma and T. Siddique, Efficacy and long-term safety of CRISPR/Cas9 genome editing in the SOD1-linked mouse models of ALS, Commun. Biol., 2021, 4, 396 CrossRef CAS PubMed.
- N. Gharaati-Far, M. R. Tohidkia, A. Dehnad and Y. Omidi, Efficiency and cytotoxicity analysis of cationic lipids-mediated gene transfection into AGS gastric cancer cells, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1001–1008 CrossRef CAS PubMed.
- A. Dousis, K. Ravichandran, E. M. Hobert, M. J. Moore and A. E. Rabideau, An engineered T7 RNA polymerase that produces mRNA free of immunostimulatory byproducts, Nat. Biotechnol., 2023, 41, 560–568 CrossRef CAS PubMed.
- S. Son, M. Park, J. Kim and K. Lee, ACE mRNA (Additional Chimeric Element incorporated IVT mRNA) for Enhancing Protein Expression by Modulating Immunogenicity, Adv. Sci., 2024, 11, e2307541 CrossRef PubMed.
- F. Ferraresso, A. W. Strilchuk, L. J. Juang, L. G. Poole, J. P. Luyendyk and C. J. Kastrup, Comparison of DLin-MC3-DMA and ALC-0315 for siRNA Delivery to Hepatocytes and Hepatic Stellate Cells, Mol. Pharm., 2022, 19, 2175–2182 CrossRef CAS PubMed.
- S. Jadhav, W. Seufert, C. Lechner and R. Schonleber, Bachem – Insights into Innovative and Sustainable Peptide Chemistry and Technology by the Leading Independent Manufacturer of TIDES, Chimia, 2021, 75, 476–479 CrossRef CAS PubMed.
- C. J. Cheng, R. Bahal, I. A. Babar, Z. Pincus, F. Barrera, C. Liu, A. Svoronos, D. T. Braddock, P. M. Glazer, D. M. Engelman, W. M. Saltzman and F. J. Slack, MicroRNA silencing for cancer therapy targeted to the tumour microenvironment, Nature, 2015, 518, 107–110 CrossRef CAS PubMed.
- V. K. Mayya and T. F. Duchaine, On the availability of microRNA-induced silencing complexes, saturation of microRNA-binding sites and stoichiometry, Nucleic Acids Res., 2015, 43, 7556–7565 CrossRef CAS PubMed.
- K. Matsumoto, K. Suzuki, H. Yasuoka, J. Hirahashi, H. Yoshida, M. Magi, M. Noguchi-Sasaki, Y. Kaneko and T. Takeuchi, Longitudinal monitoring of circulating immune cell phenotypes in anti-neutrophil cytoplasmic antibody-associated vasculitis, Autoimmun. Rev., 2023, 22, 103271 CrossRef CAS PubMed.
- E. Janik, M. Niemcewicz, M. Ceremuga, L. Krzowski, J. Saluk-Bijak and M. Bijak, Various Aspects of a Gene Editing System-CRISPR-Cas9, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- C. Happi Mbakam, G. Lamothe, G. Tremblay and J. P. Tremblay, CRISPR-Cas9 Gene Therapy for Duchenne Muscular Dystrophy, Neurotherapeutics, 2022, 19, 931–941 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, The current landscape of nucleic acid therapeutics, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- S. Soroudi, M. R. Jaafari and L. Arabi, Lipid nanoparticle (LNP) mediated mRNA delivery in cardiovascular diseases: advances in genome editing and CAR T cell therapy, J. Controlled Release, 2024, 372, 113–140 Search PubMed.
- B. P. Kleinstiver, V. Pattanayak, M. S. Prew, S. Q. Tsai, N. T. Nguyen, Z. Zheng and J. K. Joung, High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects, Nature, 2016, 529, 490–495 Search PubMed.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime, Nat. Biotechnol., 2020, 38, 824–844 Search PubMed.
- N. M. Drager, S. M. Sattler, C. T. Huang, O. M. Teter, K. Leng, S. H. Hashemi, J. Hong, G. Aviles, C. D. Clelland, L. Zhan, J. C. Udeochu, L. Kodama, A. B. Singleton, M. A. Nalls, J. Ichida, M. E. Ward, F. Faghri, L. Gan and M. Kampmann, A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states, Nat. Neurosci., 2022, 25, 1149–1162 Search PubMed.
- W. Zhou, L. Jiang, S. Liao, F. Wu, G. Yang, L. Hou, L. Liu, X. Pan, W. Jia and Y. Zhang, Vaccines’ New Era-RNA Vaccine, Viruses, 2023, 15, 1760 CrossRef CAS PubMed.
- G. An, Pharmacokinetics and Pharmacodynamics of GalNAc-Conjugated siRNAs, J. Clin. Pharmacol., 2024, 64, 45–57 Search PubMed.
- N. Bellassai, R. D'Agata and G. Spoto, Novel nucleic acid origami structures and conventional molecular beacon-based platforms: a comparison in biosensing applications, Anal. Bioanal. Chem., 2021, 413, 6063–6077 CrossRef CAS PubMed.
- S. Mueller, T. Engleitner, R. Maresch, M. Zukowska, S. Lange, T. Kaltenbacher, B. Konukiewitz, R. Ollinger, M. Zwiebel, A. Strong, H. Y. Yen, R. Banerjee, S. Louzada, B. Fu, B. Seidler, J. Gotzfried, K. Schuck, Z. Hassan, A. Arbeiter, N. Schonhuber, S. Klein, C. Veltkamp, M. Friedrich, L. Rad, M. Barenboim, C. Ziegenhain, J. Hess, O. M. Dovey, S. Eser, S. Parekh, F. Constantino-Casas, J. de la Rosa, M. I. Sierra, M. Fraga, J. Mayerle, G. Kloppel, J. Cadinanos, P. Liu, G. Vassiliou, W. Weichert, K. Steiger, W. Enard, R. M. Schmid, F. Yang, K. Unger, G. Schneider, I. Varela, A. Bradley, D. Saur and R. Rad, Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes, Nature, 2018, 554, 62–68 Search PubMed.
- J. Liu, S. Li, Q. Wang, Y. Feng, H. Xing, X. Yang, Y. Guo, Y. Guo, H. Sun, X. Liu, S. Yang, Z. Mei, Y. Zhu, Z. Cheng, S. Chen, M. Xu, W. Zhang, N. Wan, J. Wang, Y. Ma, S. Zhang, X. Luan, A. Xu, L. Li, H. Wang, X. Yang, Y. Hong, H. Xue, X. Yuan, N. Hu, X. Song, Z. Wang, X. Liu, L. Wang and Y. Liu, Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy, Blood, 2024, 143, 1825–1836 CrossRef CAS PubMed.
- Y. Gou, X. Zheng, W. Li, H. Deng and S. Qin, Polysaccharides Produced by the Mushroom Trametes robiniophila Murr Boosts the Sensitivity of Hepatoma Cells to Oxaliplatin via the miR-224-5p/ABCB1/P-gp Axis, Integr. Cancer Ther., 2022, 21, 15347354221090221 CrossRef CAS PubMed.
- I. Alekseenko, A. Kuzmich, L. Kondratyeva, S. Kondratieva, V. Pleshkan and E. Sverdlov, Step-by-Step Immune Activation for Suicide Gene Therapy Reinforcement, Int. J. Mol. Sci., 2021, 22 CAS.
- T. Oishi, M. Ito, S. Koizumi, M. Horikawa, T. Yamamoto, S. Yamagishi, T. Yamasaki, T. Sameshima, T. Suzuki, H. Sugimura, H. Namba and K. Kurozumi, Efficacy of HSV-TK/GCV system suicide gene therapy using SHED expressing modified HSV-TK against lung cancer brain metastases, Mol Ther Methods Clin Dev, 2022, 26, 253–265 CrossRef CAS PubMed.
- A. Kazlauskas, A. Darinskas, R. Meskys, A. Tamasauskas and J. Urbonavicius, Isocytosine deaminase Vcz as a novel tool for the prodrug cancer therapy, BMC Cancer, 2019, 19, 197 CrossRef PubMed.
- G. Teng, Y. Ju, Y. Yang, H. Hua, J. Chi and X. Mu, Combined antitumor activity of the nitroreductase/CB1954 suicide gene system and gamma-rays in HeLa cells in vitro, Mol. Med. Rep., 2016, 14, 5164–5170 Search PubMed.
- Y. Zhao, J. Hu, S. S. Yang, J. Zhong, J. Liu, S. Wang, Y. Jiao, F. Jiang, R. Zhai, B. Ren, H. Cong, Y. Zhu, F. Han, J. Zhang, Y. Xu, Z. Huang, S. Zhang and F. Yang, A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1, Nat. Commun., 2022, 13, 7071 Search PubMed.
- C. Wang, C. Pan, H. Yong, F. Wang, T. Bo, Y. Zhao, B. Ma, W. He and M. Li, Emerging non-viral vectors for gene delivery, J. Nanobiotechnol., 2023, 21, 272 CrossRef PubMed.
- F. Qiu, Y. Chen, C. Tang and X. Zhao, Amphiphilic peptides as novel nanomaterials: design, self-assembly and application, Int. J. Nanomed., 2018, 13, 5003–5022 CrossRef CAS PubMed.
- K. Barzaman, J. Karami, Z. Zarei, A. Hosseinzadeh, M. H. Kazemi, S. Moradi-Kalbolandi, E. Safari and L. Farahmand, Breast cancer: Biology, biomarkers, and treatments, Int. Immunopharmacol., 2020, 84, 106535 CrossRef CAS PubMed.
- X. Sun, Y. Lian, T. Tian and Z. Cui, Virus-like particle encapsulation of functional proteins: advances and applications, Theranostics, 2024, 14, 7604–7622 Search PubMed.
- H. Cui, X. Zhu, S. Li, P. Wang and J. Fang, Liver-Targeted Delivery of Oligonucleotides with N-Acetylgalactosamine Conjugation, ACS Omega, 2021, 6, 16259–16265 Search PubMed.
- Y. Tan, Y. Li, Y.-X. Qu, Y. Su, Y. Peng, Z. Zhao, T. Fu, X.-Q. Wang and W. Tan, Aptamer-Peptide Conjugates as Targeted Chemosensitizers for Breast Cancer Treatment, ACS Appl. Mater. Interfaces, 2020, 13, 9436–9444 CrossRef PubMed.
- G. Farahavar, S. S. Abolmaali, N. Gholijani and F. Nejatollahi, Antibody-guided nanomedicines as novel breakthrough therapeutic, diagnostic and theranostic tools, Biomater. Sci., 2019, 7, 4000–4016 Search PubMed.
- S. Liu, Q. Cheng, T. Wei, X. Yu, L. T. Johnson, L. Farbiak and D. J. Siegwart, Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing, Nat. Mater., 2021, 20, 701–710 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing, Nat. Nanotechnol., 2020, 15, 313–320 Search PubMed.
- J. G. Doench, N. Fusi, M. Sullender, M. Hegde, E. W. Vaimberg, K. F. Donovan, I. Smith, Z. Tothova, C. Wilen, R. Orchard, H. W. Virgin, J. Listgarten and D. E. Root, Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9, Nat. Biotechnol., 2016, 34, 184–191 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |