 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of Au-hydrides as key intermediates in the reduction of Au(III) prodrugs to active Au(I) species under protic conditions
Jasmine Ochs and
Nils Metzler-Nolte *
*
Faculty of Chemistry and Biochemistry, Inorganic Chemistry I – Bioinorganic Chemistry, Ruhr University Bochum, Universitaetsstrasse 150, Bochum 44801, Germany. E-mail: Nils.Metzler-Nolte@rub.de
First published on 7th January 2026
Abstract
Similar to PtIV prodrugs, AuIII anticancer complexes are believed to undergo intracellular reduction, thereby gaining their activity from the resulting AuI species. Unlike for PtIV, the underlying mechanism of this process remains poorly understood for AuIII. To elucidate this mechanism, we investigated the reaction of [Au(ppy)Cl2], a model AuIII complex (ppy: phenylpyridine), with two biologically relevant reductants: lipoic acid (lpa) and N-acetyl-L-cysteine-methyl ester (NAC-OMe). Our findings reveal that lpa transfers a hydride to the Au, while cysteine derivatives only bind to the metal. The Au–H complex, even visible in protic solvents by NMR spectroscopy, produced by lpa is essential for enabling a sequence of oxidative addition and reductive elimination reactions that lead to AuI species eventually. These observations provide valuable insights into the mechanisms by which anticancer gold drug candidates are reduced within the cell.
Introduction
Since cisplatin initiated the success of PtII compounds in chemotherapy, research has focused on improving the drug to minimize toxic side effects.1 An example for such an improvement is the development of PtIV prodrugs with the metal in the higher +IV oxidation state. These are slowly reduced intracellularly to their active PtII counterparts, releasing two ligands in the process.2 Numerous studies have identified sodium ascorbate, L-methionine, L-cysteine and of course glutathione (GSH) as the primary intracellular reductants.3 Sodium ascorbate, a two-electron reductant, can reduce PtII either through an inner-sphere or outer-sphere mechanism.4 In the case of GSH, it is hypothesized that the reductive elimination of the axial ligands is facilitated by the formation of a bridge between thiol and coordinated acetate.5Another approach to circumvent toxic side effects and resistances is the use of AuI and AuIII compounds, which are isoelectronic and structurally similar to the PtII/PtIV system.6 However, the high oxidation state must be stabilized for biological applications, usually using phosphine,7 N-heterocyclic carbenes,8 or polydentate ligands featuring C and N binding sites.9 Gold complexes exhibit different modes of action, such as coordination to specific enzymes like thioredoxin reductase,10 and covalent modifications of the enzyme by AuIII catalyzed cross coupling reactions,11 making them a promising new class of compounds. Beside the binding to specific targets, it is assumed that intracellular reduction by ascorbate or thiols (as for Pt) activates the AuIII complexes to their biologically active AuI analogues.12 For example, Che and co-workers employed an AuIII complex that releases its chelating ligand upon reduction, due to a change to linear geometry. Thereby, the fluorescent ligand is activated and the cytotoxic AuI-NHC complex bearing a GSH ligand is formed.13 Another possibility is the release of free AuI ions from polypyridyl AuIII complexes, which leads to inhibition of thioredoxin reductase (TrxR), even under hypoxic conditions. Additionally, the ligand exhibits a second mode of action by binding to potassium channels, thereby damaging the lysosomal membrane.14 Interestingly, almost nothing is known about the actual reduction pathway, but cysteine and especially GSH are assumed to facilitate the reduction. Recent work by Mironov and Kharlamova15 on N^N bidentate Au complexes suggests ligand substitution, followed by “inner-sphere reduction” and the formation of polymeric AuI-thiolate species as a possible pathway. However, no intermediates could be isolated in this work, and the exact mechanism of reduction remains elusive. The analyzed N^N gold complexes are very prone to reduction, also rapidly leading to ligand loss. Notably, in these cases, the anticancer activity of such complexes is attributed to the ligand itself rather than the Au center.12,16 In contrast, highly stabilized AuIII complexes bearing bidentate C^N ligands bridged by CO, CH2, or NH groups also undergo first a ligand exchange but afterwards, in the presence of a sulphur or selenium containing amino acid, a C–S/Se cross coupling with the C^N ligand was observed, releasing free AuI.17 However, this process seems to be specific for the ligand type used in this work, and no intermediates were identified.
To gain a more general overview of the mechanism of reduction, we present here a study on a stabilized AuIII model complex with two naturally relevant reductants. We selected AuIII-dichlorido-phenylpyridine ([Au(ppy)Cl2]) (see Scheme 1 for its chemical formula) as a member of the cytotoxic bidentate C^N complexes, known for its stable +III oxidation state.18 For the biologically occurring reductants (see Fig. 1), we used N-acetyl-L-cysteine methyl ester (NAC-OMe) as a simplified analogue of GSH, and lipoic acid (lpa). Lipoic acid, like GSH, is an organosulfur compound, but it contains two thiol groups capable of forming a five-membered ring.19 Its unique structure facilitates substrate channeling between various active sites in multienzyme complexes, making it an essential cofactor for the citric acid cycle.20 Additionally, its low redox potential makes it a powerful reductant, capable of scavenging free reactive oxygen species (ROS). This dual functionality significantly contributes to maintaining a balanced redox system intracellularly.21 Using this model system, we were able to gain a deeper understanding of the reduction mechanism of AuIII complexes.
 | ||
| Fig. 1 Structure of lipoic acid in the cyclic, oxidized form (left), the reduced species with two thiols (middle), and N-acetyl-L-cysteine methyl ester (NAC-OMe) (right). | ||
Results and discussion
Reaction with lipoic acid
First, we utilized naturally occurring lpa, known for its antioxidant properties. To address the poor water solubility of the Au precursor, reactions were performed in a DCM/MeOH (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture. A further advantage of this solvent is the slower reaction rate compared to water, which facilitates the analysis of intermediates. Additionally, the solvent was degassed to prevent the oxidation of the dithiol to the corresponding disulfide.
1) mixture. A further advantage of this solvent is the slower reaction rate compared to water, which facilitates the analysis of intermediates. Additionally, the solvent was degassed to prevent the oxidation of the dithiol to the corresponding disulfide.
The reaction between [Au(ppy)Cl2] and reduced lpa resulted in the formation of gold nanoparticles, visible as purple colloids in the mixture, as well as elemental gold as a side product, indicating the potential of lpa for AuIII reduction. Furthermore, various molecular Au compounds were detected as intermediates. After silica column purification of various batches, three compounds were isolated and analyzed by NMR spectroscopy and mass spectrometry.
The most often observed complex is [AuH(ppy)(lpa)2], which has a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry of ppy to lpa, as evident in the 1H NMR spectrum (see Fig. 2). The 1H NMR spectrum reveals the presence of two non-coordinating thiols, appearing as a multiplet at 1.47 ppm, which were further confirmed using Ellmann's reagent. Additionally, the singlet at 3.67 ppm indicates the esterification of lpa by methanol. The required HCl is released during lpa binding to gold, and because the reaction is carried out in a DCM/methanol mixture, it is non stabilized and, consequently, highly reactive. However, this reaction is not physiologically relevant, but rather an artifact caused by the presence of methanol in the solvent mixture, which is necessary for precursor solubility. An (unexpected) additional aromatic proton signal is observed at 8.03 ppm, showing the breakage and protonation of the Au–C bond, while ppy is still coordinated through the nitrogen atom. The fourth binding site is not any more occupied by a chlorido ligand, as evidenced by a negative “Purple of Cassius” test. This test is specific for AuIII-chlorides, which are reduced by SnII chloride to form a purple pigment. Instead, the last ligand is an Au hydride, visible as a broad signal at 2.78 ppm. This hypothesis is supported by data from the Bochmann group which have isolated and confirmed the existence of AuIII-hydrides with bi- and tridentate C- and N-cyclometallating ligands.22,23 They demonstrated that such hydrides occur in the 1H NMR spectra over a wide range from −8.5 ppm up to 7.0 ppm, which aligns well with our findings. All other data (including 2D COSY-NMR, see SI) confirm the proposed structure of [AuH(ppy)(lpa)2] as shown in Fig. 3.
2 stoichiometry of ppy to lpa, as evident in the 1H NMR spectrum (see Fig. 2). The 1H NMR spectrum reveals the presence of two non-coordinating thiols, appearing as a multiplet at 1.47 ppm, which were further confirmed using Ellmann's reagent. Additionally, the singlet at 3.67 ppm indicates the esterification of lpa by methanol. The required HCl is released during lpa binding to gold, and because the reaction is carried out in a DCM/methanol mixture, it is non stabilized and, consequently, highly reactive. However, this reaction is not physiologically relevant, but rather an artifact caused by the presence of methanol in the solvent mixture, which is necessary for precursor solubility. An (unexpected) additional aromatic proton signal is observed at 8.03 ppm, showing the breakage and protonation of the Au–C bond, while ppy is still coordinated through the nitrogen atom. The fourth binding site is not any more occupied by a chlorido ligand, as evidenced by a negative “Purple of Cassius” test. This test is specific for AuIII-chlorides, which are reduced by SnII chloride to form a purple pigment. Instead, the last ligand is an Au hydride, visible as a broad signal at 2.78 ppm. This hypothesis is supported by data from the Bochmann group which have isolated and confirmed the existence of AuIII-hydrides with bi- and tridentate C- and N-cyclometallating ligands.22,23 They demonstrated that such hydrides occur in the 1H NMR spectra over a wide range from −8.5 ppm up to 7.0 ppm, which aligns well with our findings. All other data (including 2D COSY-NMR, see SI) confirm the proposed structure of [AuH(ppy)(lpa)2] as shown in Fig. 3.
 | ||
| Fig. 3 All observed gold complexes from the reaction of lpared and [Au(ppy)Cl2] with color code for NMR spectra analysis. | ||
The second isolated complex seems to be an intermediate structure, which is quite similar to [AuH(ppy)(lpa)2]. The striking difference between both complexes is the ratio of ppy and lpa as detected in the 1H NMR spectrum (see SI). The ratio of two ppy to three lpa ligands clearly shows the formation of the bridged binuclear complex [Au2H2(ppy)2(lpa)3], as depicted in Fig. 3 (middle). Apart from the altered stoichiometry, the NMR spectrum exhibits the same features, including the Au-H signal. Additionally, the structure was further confirmed by a signal in the ESI-MS at m/z = 685.2, which corresponds to the doubly protonated species.
Interestingly, a third intermediate was identified experimentally as an AuI rather than an AuIII compound. The ratio of ppy to lpa is altered again, this time with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, resulting in the complex [Au(ppy)(lpa)] as depicted in Fig. 3. Unlike the other two complexes, no hydride signal was detected, and only one thiol proton was observed. Furthermore, the pyridyl-proton signals are downfield-shifted and overlap with the phenyl ring signals, indicating a geometry change from the square-planar AuIII to the linear AuI complex. The formation of this AuI intermediate clearly highlights the reduction and oxidation processes which lead to the formation of such complexes.
1 ratio, resulting in the complex [Au(ppy)(lpa)] as depicted in Fig. 3. Unlike the other two complexes, no hydride signal was detected, and only one thiol proton was observed. Furthermore, the pyridyl-proton signals are downfield-shifted and overlap with the phenyl ring signals, indicating a geometry change from the square-planar AuIII to the linear AuI complex. The formation of this AuI intermediate clearly highlights the reduction and oxidation processes which lead to the formation of such complexes.
Investigation of the reduction mechanism with lipoic acid
To shed further light on the mechanism, additional experiments were conducted. First, the reaction was performed in deuterated solvents to establish the origin of the new aromatic proton, the methyl group, and the hydride from the thiols or solvent. For this purpose, the crude reaction mixture was directly measured to investigate changes compared to the previous NMR spectra. Additionally, a 2D NMR spectrum was recorded after switching to non-deuterated solvents. The spectral comparison is depicted Fig. 4 for [Au2H2(ppy)2(lpa)3]. | ||
| Fig. 4 NMR spectra comparison of the reaction in non-deuterated (exemplarily for [Au2H2(ppy)2(lpa)3]) and deuterated solvents as well as the corresponding 2D NMR spectrum. The signals are color-coded corresponding to Fig. 3 with the asterisk marking the occurrence of the ppy-H, Au-H, thiol and methyl ester signals either the 1H or 2D NMR spectrum. For the aromatic region, the number of protons per signal is given. | ||
Performing the reaction in deuterated solvents reduces the number of aromatic protons to 16, in turn the signal for the new ppy-D appears at 8.12 ppm in the 2D NMR spectrum. Additionally, the methyl group at 3.64 ppm disappears in the 1H NMR spectrum of the reaction with deuterated solvents and can instead be found in the 2D NMR. Interestingly, the Au-D signal is present in the 2D NMR at 2.83 ppm, while the thiol signals were found at 1.45 ppm in the 1H NMR. This indicates that the metal hydride bond is more stable than the S–H bond towards H/D exchange even in protic solvents. These results support the assumption of lpa esterification with methanol and confirm the participation of CD3OD or the thiol after H/D exchange in the Au–C bond cleavage and hydride formation.
To identify the initial reaction steps, the reaction time was decreased from 16 hours to 3 hours. During this time, the [Au(ppy)Cl2] must have reacted as it was completely dissolved. Interestingly, only lpaox was found but no gold lpa complex.
Combining all the results, the first part of the reaction mechanism is proposed in Scheme 1. The reaction starts with ligand exchange of a chloride to a thiolate of dihydro lipoic acid and the simultaneous liberation of hydrogen chloride (see Scheme 1(i)). By comparison with the cysteine (Cys) containing gold complex [Au(ppy)(Cys)](NO3) reported in the literature,24 this ligand exchange is assumed to occur in the trans position to the pyridyl nitrogen.
Expected from the work previously done in our group, the next step would be the deprotonation of the second thiol with subsequent metal coordination.25 However, in the absence of the peptide backbone, lpa oxidizes to the thermodynamically stable five-membered ring, which was confirmed as the disulfide form was the only detected lpa species after three hours. Therefore, the released hydride is then transferred to the AuIII complex, substituting the thiolate as the negatively charged ligand. Bochmann et al. showed that in binuclear bonded AuIII hydride complexes, the hydride binds trans to the carbonyl–gold bond.22 Consequently, we believe that in our reaction the trans position to the Au–C bond should also be favored (Scheme 1(ii)).
From complex 2, the C–H bond cannot be reductively eliminated as the hydride and carbanion are spatially separated. This strongly suggests that the Au–H is donated by lpa-H2. The formed hydride complex 2 is reactive, but the reverse reaction back to complex 1 is disfavored because the stable five-membered ring in oxidized lpa would have to be opened up again.
However, AuI complexes have been reported to undergo oxidative addition with C–C bonds26 and even with the higher analogue Si–Si bonds,27 using conditions similar to those in this work. Even more strikingly, Gray and co-workers showed that the oxidative addition of a disulfide to AuI and the reductive elimination of two thiols from AuIII are reversible processes for their studied example.28
To facilitate the reduction of the complex 2, a reductive elimination is necessary. However, the trans positioning of the carbanion and hydride impedes this reaction. Consequently, a ligand exchange must occur, replacing the chloride with a second hydride, which can be donated by lpared or methanol. lpared was employed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with the [Au(ppy)Cl2] complex and is presumably consumed during the first reaction step, leaving only a small amount of hydride. On the other hand, methanol oxidation to formaldehyde could also occur. Although methanol is an extremely mild reductant, alcohols are well-established reductants for metal complexes and its interactions with AuIII could facilitate the reaction. In a cellular environment, however, the hydride donor is more likely lpa, NAD(P)H, or a similar biological reductant.
1 ratio with the [Au(ppy)Cl2] complex and is presumably consumed during the first reaction step, leaving only a small amount of hydride. On the other hand, methanol oxidation to formaldehyde could also occur. Although methanol is an extremely mild reductant, alcohols are well-established reductants for metal complexes and its interactions with AuIII could facilitate the reaction. In a cellular environment, however, the hydride donor is more likely lpa, NAD(P)H, or a similar biological reductant.
As a result of the hydride transfer, complex 3 then has a hydride in the cis position to the carbanion, enabling reductive elimination with C–H bond formation. Such C–H bond formations through oxidative addition and reductive elimination reactions are well-established processes with gold complexes as catalysts.22,29 This process is further favored because the hydride opposite the Au–C bond exerts a strong trans-effect, resulting in the formation of 5, which is a linear AuI species. This complex is now able to undergo oxidative addition with the oxidized cyclic lipoic acid, forming a complex chelated by both thiolates of lpa (complex 6). It is most likely that esterification of the carboxylic acid (Scheme 1(iii)) occurred before addition to 5. The HCl produced by the ligand exchange reaction is highly reactive and, in the DCM/MeOH mixture, is strong enough to catalyze the esterification with methanol at room temperature. As a result, the methanol ester of lipoic acid 4 reacts with the AuI complex 5.
With this mechanism, the occurrence of the unexpected additional protons of the ppy ligand, the Au–H, and the ester signal can be explained. However, the observed complexes are still not completely explained by this mechanism. To monitor further changes, NMR spectra were recorded at each step in the synthesis and purification process: before solvent evaporation, after solvent evaporation, and after silica column chromatography. As shown in Fig. 5, the formation of 6 in the crude reaction mixture is clearly proven. However, solvent evaporation at 40 °C alters the complex structure, and only after the purification the product NMR spectrum is obtained. Hence, complex 6 undergoes further changes during purification.
 | ||
| Fig. 5 NMR spectra of the reaction in deuterated solvents directly after the 16 h reaction (top), after solvent evaporation (middle) and purification by filtration over a silica column (bottom). | ||
Formation of polynuclear species
For this reason, we propose a mechanism for the second part (see Scheme 2) towards the characterized products, which is initiated by either heat or interaction with the silica column material. The following reaction steps are a cascade of different reductive elimination and oxidative addition steps, which eventually explain the formation of all the observed intermediates shown in Fig. 3.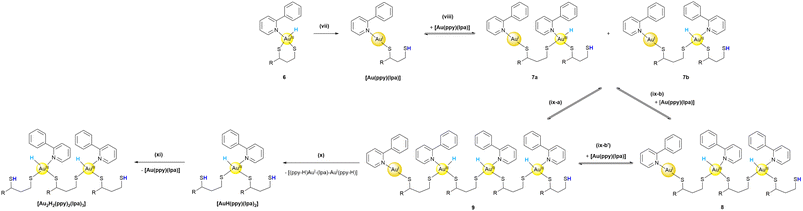 | ||
| Scheme 2 Proposed mechanism (part II) for the formation of the obtained products [Au(ppy)(lpa)], [Au2H2(ppy)2(lpa)3] and [AuH(ppy)(lpa)2] via reductive elimination and oxidative addition reactions, colour coding as in Fig. 1. (vii) Reductive elimination of the S–H bond initiated during purification, (viii) dimerization of [Au(ppy)(lpa)] by oxidative addition of S–H in a non-concerted (7a) or concerted (7b) fashion, (ix-a) direct oxidative addition of 7a and 7b or (ix-b) and (ix-b′) two times oxidative addition of [Au(ppy)(lpa)], (x) reductive elimination of [(ppy-H)AuI-(lpa)-AuI(ppy-H)] towards [Au2H2(ppy)2(lpa)3], followed by reductive elimination (xi) of [Au(ppy)(lpa)] yielding [AuH(ppy)(lpa)2]. | ||
First, one thiol (RS-H) is reductively eliminated from 6, resulting in the isolated complex [Au(ppy)(lpa)] (Scheme 2(vii)). This aligns with the findings of Bochmann et al., who reported that AuI species originating from AuIII hydrides by C–H elimination can be isolated.22 Assuming that the reductive elimination of S–H is a reversible process, the oxidative addition of an S–H bond to a second molecule of [Au(ppy)(lpa)] is possible (step (viii)). Again, a reaction with 4 is not reasonable as an equimolar ratio of [Au(ppy)Cl2] and lpared was used and the lipoic acid derivatives are assumed to be fully converted during each reaction step. The addition can take place in a non-concerted (ix-a) or concerted (ix-b) fashion. Further oxidative additions of free thiols to other AuI-complexes lead to polymerization (see steps (ix)). Polymerization is a process often observed under acidic conditions for AuI species, but usually led to Au–S–Au bridged complexes when monodentate ligands were used.30
In contrast to synthesis procedures for AuIII-hydride complexes in the literature, the reactions in this work were performed at room temperature rather than at low temperatures. For the previously reported complexes, rearrangements and reductive eliminations are favored above 228 K.22 Consequently, ligand rearrangements are probable for polymeric structures like 9. Positional changes of the monodentate ligands, the hydride and ppy are likely. After the rearrangement, reductive elimination is not only possible as a reverse reaction, but also a new complex can be formed. Step (x) shows the chain breakdown that decreases the length of the polymeric structure, which is entropically favored as more and smaller molecules are obtained. This results in the experimentally obtained [Au2H2(ppy)2(lpa)3] complex, which can subsequently undergo a final reductive elimination of an S–H bond yielding [AuH(ppy)(lpa)2], which was isolated and characterized.
The proposed mechanism explains all 1H-NMR signals for all detected complexes. Moreover, it aligns with the experimental observation that [Au(ppy)(lpa)] was only observed once, [Au2H2(ppy)2(lpa)3] several times and [AuH(ppy)(lpa)2] was obtained predominantly as the most stable product. Nonetheless, all compounds are still very unstable and cannot be stored in solution or dried. Consequently, additional characterization methods, such as elemental analysis and X-ray single crystal structures, could not be employed.
Reaction with cysteine derivatives
To determine whether lipoic acid possesses unique properties in the reduction of AuIII through the formation of the five-membered ring, N-acetyl-L-cysteine-methyl ester (NAC-OMe) was employed as a second naturally relevant reductant. The protection of both sites was selected to mimic the properties of glutathione (GSH) and to minimize amine coordination.The reaction was carried out similarly to the lipoic acid experiments, but with either one or two equivalents of NAC-OMe. One equivalent corresponds to the same amount of ligand, while two equivalents equate to the number of reduction equivalents in lpa. The reaction mixtures were analysed directly without purification.
Unexpectedly, unreacted NAC-OMe was found in both reaction mixtures. This contrasts with the crude NMR of the lipoic acid reaction and suggests a different reaction mechanism. The formed complex and the ligand were distinguished and assigned using a DOSY spectrum. The 1H NMR spectrum (see Fig. 6) reveals an intact Au–C bond, indicated by the presence of eight aromatic proton signals. NAC-OMe and ppy were found in the same ratio, which was for the lpa complexes an exceptional case for the AuI complex 5.
The NMR analysis allows two possible binding modes, depicted in Fig. 6. The complex could be similar to 2, with cysteine acting as a monodentate ligand binding through the thiolate (11). Alternatively, it could bind in a bidentate N^S fashion (10). This would align with the structures published by Konno et al.24 and Lee et al.,31 where they synthesised mononuclear AuIII complexes in aqueous solutions with cysteine derivatives acting as chelating ligands. The signal in ESI-MS at m = 585.4 m/z matches the mass of [11 + Na]+.
Moreover, single crystals of the square-planar gold complex 11 were obtained after several days, indicating greater stability compared to the lpa complexes. The crystal structure also supports the monodentate binding mode (Fig. 7A). Originating from the bidentate ppy ligand, which forms a five-membered ring with Au, the square planar structure is slightly distorted. The C–Au–N angle is only 81.38°, while the S–Au–Cl angle is extended to 96.40°. The trans-effect is evident, as the Au–C bond is shortened to 2.062 Å, whereas the Au–Cl bond is elongated to 2.377 Å. The position of the thiolate trans to the pyridyl-N aligns with the proposed first step in the binding cascade of lipoic acid, further supporting the mechanism (step (i) in Scheme 1). Additionally, these results show that C–S cross coupling using AuIII complexes requires bridged C^N ligands,17 and is not a general mode of action.
 | ||
| Fig. 7 ORTEP-plot of 11 (A) and [AuCl(ppy)(HC–OMe)] (B). Ellipsoids are drawn at the 50% probability level. For [AuCl(ppy)(HC–OMe)], the solvent and counter ion are omitted for clarity. | ||
Additionally, single crystals with cysteine-methyl ester (HC-OMe) were grown to investigate whether the binding difference arises from the synthesis conditions or the less electron-rich acetyl-protected nitrogen. The crystal structure displays the same monodentate binding mode as with NAC-OMe, with matching square-planar geometry, bond lengths, and angles (Fig. 7B). In contrast to its acetyl-protected analogue, the free amine is protonated due to the presence of HCl released during the reaction. The resulting positive charge prevents bidentate coordination, which was observed in aqueous media, and instead promotes monodentate binding. This highlights the different possibilities of cysteine–gold binding, which are strongly influenced by the solvent environment.
Remarkably, all cysteine derivatives bind to the gold metal but only lpa is able to reduce AuIII to AuI. The redox properties of lpa and the essential antioxidant GSH were measured elsewhere at 25 °C and pH 7, yielding standard reduction potentials of E0 = −0.29 V and E0 = −0.23 V, respectively.32 These values demonstrate that lpa is the stronger reductant33 and is capable of reducing AuIII, whereas GSH or its derivatives primarily act as ligands, binding to the gold complex without reducing it.
It should be noted that this work was performed under degassed conditions/N2 protection to ensure a high concentration of reduced lpa at all times. However, the presence of oxygen in biological systems is not assumed to change our conclusions as lpa can then be constantly reduced by the cellular machinery.
Experimental
Materials and methods
All chemicals and solvents were purchased from commercial suppliers and used without further purification. Solvents were degassed with nitrogen for 10 min prior to use or used as received. 1H NMR and 13C{1H} NMR measurements were conducted at 298 K on a Bruker AV-III 300, and residual solvent signals from the deuterated solvents were used to reference the 1H or 13C signals. MestReNova was used to perform spectrum analysis, signals were reported in parts per million (ppm) with coupling constants (J) given in Hz. The numbering scheme for signal assignment is found in the SI. Electron spray ionization mass spectra were recorded using a Bruker Esquire 6000 in the positive sensitivity mode. For single-crystal diffraction studies, a Synergy S dual S3 wavelength diffractometer employing mirror monochromatic CuKα radiation generated from a PhotonJet-S (Cu) X-ray Source with a HyPix-600 HE detector was used for data collection. Cell constants were obtained from a least square's refinement. CrysAlisPro was used for integration, reduction, and absorption correction of the data, measured at 100.15 K. The structures were solved using SHELXT 2018/2 (ref. 34) and refined using SHELXL 2018/3.35 The Olex2 1.3 (ref. 36) interface was used to generate publication material.Synthetic procedures
[Au(ppy)Cl2] was synthesised according to the literature procedure.37 Tetrachloroauric acid (0.05 g, 0.13 mmol, 1.0 eq.) was dissolved in 4 mL water, and 2-phenylpyridine (0.02 mL, 0.14 mmol, 1.1 eq.) was added. Immediately, some solid precipitated. The solution was irradiated for one hour in a microwave (55 W, 170 °C, 13 bar). The formed solid was filtered off, washed with water (3 × 2 mL) and dried for 1 h under a N2 stream. The product was obtained as a light-yellow solid. Yield: 0.03 g (0.08 mmol, 63%). 1H NMR (300 MHz, DMSO-d6): δ (ppm) 9.53 (1H, d, HA, JHA−HB = 6.10 Hz), 8.42 (2H, m, 1xHC, 1xHD), 7.98 (1H, d, HE, JHE−HF = 7.52 Hz), 7.82 (1H, d, HH), 7.77 (1H, td, HB, JHB−HA = 6.20 Hz), 7.48 (1H, t, HF, JHF−HE = 7.47 Hz), 7.38 (1H, t, HG).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and tris(2-carboxy ethyl)phosphine hydrochloride (TCEP) (0.28 g, 0.97 mmol, 2.0 eq.) was dissolved in 3 mL MeCN/H2O (3
1) and tris(2-carboxy ethyl)phosphine hydrochloride (TCEP) (0.28 g, 0.97 mmol, 2.0 eq.) was dissolved in 3 mL MeCN/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Both solutions were degassed with N2 for 10 min. TCEP was added to the yellow lpa solution, which was decolorized within 5 min. The mixture was stirred for one hour at room temperature, and the aqueous phase was extracted with DCM (3 × 10 mL). After drying the combined organic phases over MgSO4, the solvent was removed under reduced pressure, resulting in a colorless liquid. Yield: 0.08 g (0.37 mmol, 77%). 1H NMR (300 MHz, CDCl3): δ (ppm) 10.98 (1H, s, COOH), 2.96–2.85 (1H, m, Hg), 2.78–2.58 (2H, m, Hi), 2.36 (2H, t, Hc), 1.94–1.83 (1H, m, Hh), 1.78–1.71 (1H, m, Hh), 1.69–1.39 (6H, m, 2xHd, 2xHe, 2xHf), 1.34 (1H, t, SHi), 1.30–1.28 (1H, d, SHg). 13C{1H} NMR (75 MHz, CDCl3): δ (ppm) 179.6 (C
1). Both solutions were degassed with N2 for 10 min. TCEP was added to the yellow lpa solution, which was decolorized within 5 min. The mixture was stirred for one hour at room temperature, and the aqueous phase was extracted with DCM (3 × 10 mL). After drying the combined organic phases over MgSO4, the solvent was removed under reduced pressure, resulting in a colorless liquid. Yield: 0.08 g (0.37 mmol, 77%). 1H NMR (300 MHz, CDCl3): δ (ppm) 10.98 (1H, s, COOH), 2.96–2.85 (1H, m, Hg), 2.78–2.58 (2H, m, Hi), 2.36 (2H, t, Hc), 1.94–1.83 (1H, m, Hh), 1.78–1.71 (1H, m, Hh), 1.69–1.39 (6H, m, 2xHd, 2xHe, 2xHf), 1.34 (1H, t, SHi), 1.30–1.28 (1H, d, SHg). 13C{1H} NMR (75 MHz, CDCl3): δ (ppm) 179.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 42.9 (Cc), 39.4 (CSH), 38.9 (CSH), 34.0 (Ch), 26.6 (Caliph), 24.4 (Caliph), 22.4 (Caliph).
O), 42.9 (Cc), 39.4 (CSH), 38.9 (CSH), 34.0 (Ch), 26.6 (Caliph), 24.4 (Caliph), 22.4 (Caliph).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and degassed with N2 for 10 min, to maintain the reduced form of lpa in the subsequent reaction. A solution of dihydro lipoic acid (0.01 g, 0.05 mmol, 1.0 eq.) in degassed DCM/MeOH (9
1) and degassed with N2 for 10 min, to maintain the reduced form of lpa in the subsequent reaction. A solution of dihydro lipoic acid (0.01 g, 0.05 mmol, 1.0 eq.) in degassed DCM/MeOH (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added slowly, resulting in an immediate change from colorless to yellow. After stirring the solution under N2 for 16 h at room temperature, the solvent was removed under reduced pressure. The solid was purified by column chromatography on silica (10 mL), eluting with n-hexane/EtOAc. The product was obtained as a yellow-brownish oil after removing the solvent under reduced pressure.
1) was added slowly, resulting in an immediate change from colorless to yellow. After stirring the solution under N2 for 16 h at room temperature, the solvent was removed under reduced pressure. The solid was purified by column chromatography on silica (10 mL), eluting with n-hexane/EtOAc. The product was obtained as a yellow-brownish oil after removing the solvent under reduced pressure.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.83 (1H, d, HA), 8.08 (2H, m, 1xHE, 1xHM), 8.02 (1H, d, HC), 7.89 (1H, d, HD), 7.59–7.46 (4H, m, 1xHF, 1xHG, 1xHH, 1xHB), 3.67 (3H, s, COOMe) 3.57 (1H, q, Hg), 3.23–3.07 (2H, m, Hi), 2.46 (1H, sex, Hh), 2.33 (2H, t, Hc), 1.91 (1H, sex, Hh), 1.74–1.61 (4H, m, 2xHd, 2xHf), 1.52–1.40 (3H, m, 1xSH, 2xHe).
1). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.83 (1H, d, HA), 8.08 (2H, m, 1xHE, 1xHM), 8.02 (1H, d, HC), 7.89 (1H, d, HD), 7.59–7.46 (4H, m, 1xHF, 1xHG, 1xHH, 1xHB), 3.67 (3H, s, COOMe) 3.57 (1H, q, Hg), 3.23–3.07 (2H, m, Hi), 2.46 (1H, sex, Hh), 2.33 (2H, t, Hc), 1.91 (1H, sex, Hh), 1.74–1.61 (4H, m, 2xHd, 2xHf), 1.52–1.40 (3H, m, 1xSH, 2xHe).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.79 (2H, d, HA), 8.06 (4H, d, 2xHE, 2xHM), 7.95 (2H, t, HC), 7.84 (2H, d, HD), 7.56–7.48 (6H, m, 2xHF, 2xHG, 2xHH), 7.40 (2H, t, HB), 3.67 (9H, s, COOMe), 3.57 (3H, q, Hg), 3.23–3.07 (6H, m, Hi), 2.78 (2H, s, Au–H), 2.46 (3H, sex, Hh), 2.33 (6H, t, Hc), 1.90 (3H, sex, Hh), 1.74–1.61 (12H, m, 6xHd, 6xHe), 1.52–1.42 (8H, m, 6xHf, 2x SH). ESI-MS (pos): m/2z 685.2, calc. for [C49H70Au2N2O6S6]2+ = 685.2.
1). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.79 (2H, d, HA), 8.06 (4H, d, 2xHE, 2xHM), 7.95 (2H, t, HC), 7.84 (2H, d, HD), 7.56–7.48 (6H, m, 2xHF, 2xHG, 2xHH), 7.40 (2H, t, HB), 3.67 (9H, s, COOMe), 3.57 (3H, q, Hg), 3.23–3.07 (6H, m, Hi), 2.78 (2H, s, Au–H), 2.46 (3H, sex, Hh), 2.33 (6H, t, Hc), 1.90 (3H, sex, Hh), 1.74–1.61 (12H, m, 6xHd, 6xHe), 1.52–1.42 (8H, m, 6xHf, 2x SH). ESI-MS (pos): m/2z 685.2, calc. for [C49H70Au2N2O6S6]2+ = 685.2.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.76 (1H, d, HA), 8.03 (2H, d, 1xHE, 1xHM), 7.89 (1H, t, HC), 7.81 (1H, d, HD), 7.53–7.44 (3H, m, 1xHF, 1xHG, 1xHH), 7.35 (2H, t, HB), 3.67 (6H, s, COOMe), 3.57 (2H, quin, Hg), 3.22–3.06 (4H, m, Hi), 2.78 (1H, s, Au–H), 2.45 (2H, sex, Hh), 2.32 (4H, t, Hc), 1.92 (2H, sex, Hh), 1.73–1.61 (8H, m, 4xHd, 4xHf), 1.57–1.36 (6H, m, 2xSH, 4xHe). 13C{1H} NMR (75 MHz, CDCl3): δ (ppm) 174.1 (C
1). 1H NMR (300 MHz, CDCl3): δ (ppm) 8.76 (1H, d, HA), 8.03 (2H, d, 1xHE, 1xHM), 7.89 (1H, t, HC), 7.81 (1H, d, HD), 7.53–7.44 (3H, m, 1xHF, 1xHG, 1xHH), 7.35 (2H, t, HB), 3.67 (6H, s, COOMe), 3.57 (2H, quin, Hg), 3.22–3.06 (4H, m, Hi), 2.78 (1H, s, Au–H), 2.45 (2H, sex, Hh), 2.32 (4H, t, Hc), 1.92 (2H, sex, Hh), 1.73–1.61 (8H, m, 4xHd, 4xHf), 1.57–1.36 (6H, m, 2xSH, 4xHe). 13C{1H} NMR (75 MHz, CDCl3): δ (ppm) 174.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 129.3 (Carom), 127.5 (Carom), 122.9 (Carom), 56.5 (Cc), 51.7 (Cb), 40.4 (CSH), 38.6 (CSH), 34.7 (Caliph), 34.0 (Caliph), 28.9 (Caliph), 24.8 (Caliph).
O), 129.3 (Carom), 127.5 (Carom), 122.9 (Carom), 56.5 (Cc), 51.7 (Cb), 40.4 (CSH), 38.6 (CSH), 34.7 (Caliph), 34.0 (Caliph), 28.9 (Caliph), 24.8 (Caliph).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and degassed with N2 for 10 min. A solution of cysteine derivative (12.6 mg, 0.07 mmol, 2.0 eq. or 6.3 mg, 0.04 mmol, 1.0 eq.) in 0.5 mL degassed DCM/MeOH (9
1) and degassed with N2 for 10 min. A solution of cysteine derivative (12.6 mg, 0.07 mmol, 2.0 eq. or 6.3 mg, 0.04 mmol, 1.0 eq.) in 0.5 mL degassed DCM/MeOH (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added slowly, resulting in a color change from colorless to yellow within 5 minutes. After stirring the solution under N2 for 17 h at room temperature, the solvent was removed under reduced pressure. NMR spectra of the crude yellow products were measured without purification.
1) was added slowly, resulting in a color change from colorless to yellow within 5 minutes. After stirring the solution under N2 for 17 h at room temperature, the solvent was removed under reduced pressure. NMR spectra of the crude yellow products were measured without purification.Conclusions
Beyond the almost ubiquitous Pt anticancer compounds, a few Au complexes in the oxidation state +I are in clinical use. In addition, also AuIII complexes were suggested as promising drug candidates. It is generally assumed that intracellular reduction activates the AuIII complexes to their biologically active AuI analogues, however details remain elusive. Herein, we demonstrate a pathway for the reduction of AuIII to AuI complexes using a biological relevant reductant, by isolation and characterization of several intermediates by NMR spectroscopy and MS. The reaction mechanism involves AuIII hydrides, and a series of oxidative addition and reductive elimination reactions occurs. Noteworthy, Au hydrides (which were previously generated only under rigorously dry conditions at temperatures below 0 °C)22 were observed in this work in a protic solvent at room temperature.Essential for the gold reduction is the use of naturally abundant lipoic acid (lpa) instead of GSH derivatives on our tested system. It is noteworthy that the reaction with simple thiols, such as cysteine methyl ester and its N-acetylated analogue (NAC-OMe), does not yield the same reduction products under identical conditions. While in all cases, thiol coordination on the AuIII center is the first step, only lpa will induce further reduction of the Au center through formation of Au hydrides. This suggests that the key factor is the slightly more negative reduction potential of lpa,32,33 and possibly also the entropically favoured formation of cyclic products in the case of lpa only. Therefore, we recommend that lpa should be considered in stability tests involving redox-sensitive complexes, to ensure that the observed activity is correctly attributed to the appropriate complex and metal oxidation state. Additionally in this work, the formation of di- and polymeric Au-clusters suggests a route for intracellular AuIII reduction, which could be the first steps towards the formation of gold nano particles – a side reaction that is often observed in gold chemistry.
Finally, our work bears important implications for the future design of AuIII complexes in medicinal inorganic chemistry. Previous work has already suggested that AuIII compounds are prodrugs which are converted into the active species by an activation-by-reduction mechanism. With a more detailed, mechanistic view from this work on the exact reduction mechanism and Au hydride species as possible intermediates, it is now possible to design AuIII prodrugs in a more rational way. Also, the importance of matching redox potentials in Au complexes to biological reductants becomes apparent. And lastly, a paradigm arises for the design of redox-inert AuIII complexes. In such compounds, hydride formation and particularly reductive elimination from any possible Au–H intermediates should be prevented by ligand design.
Author contributions
J. O.: investigation, formal analysis, writing – original draft; N. M.-N.: conceptualization, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
CCDC 2424404(11) and 2424406(12) contain the supplementary crystallographic data for this paper.38a,bAll other supporting data to this article is included in the Supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc06212h.
Acknowledgements
This study was supported by the German Research Foundation (DFG; Research Training Group GRK2341 “Microbial Substrate Conversion (MiCon)”).Notes and references
- L. Kelland, Nat. Rev. Cancer, 2007, 7, 573–584 Search PubMed.
- (a) T. C. Johnstone, K. Suntharalingam and S. J. Lippard, Chem. Rev., 2016, 116, 3436–3486 Search PubMed; (b) J. X. Ong, C. S. Q. Lim, H. van Le and W. H. Ang, Angew. Chem., Int. Ed. Engl., 2019, 58, 164–167 Search PubMed; (c) S. Yuan, Y. Zhu, Y. Dai, Y. Wang, D. Jin, M. Liu, L. Tang, F. Arnesano, G. Natile and Y. Liu, Angew. Chem., Int. Ed. Engl., 2022, 61, e202114250 CrossRef CAS PubMed.
- (a) E. Wexselblatt and D. Gibson, J. Inorg. Biochem., 2012, 117, 220–229 CrossRef CAS PubMed; (b) S. Chen, Q. Zhou, K.-Y. Ng, Z. Xu, W. Xu and G. Zhu, Inorg. Chem. Front., 2024, 11, 3085–3118 Search PubMed.
- (a) Z. Xu, Z. Wang, Z. Deng and G. Zhu, Coord. Chem. Rev., 2021, 442, 213991 CrossRef CAS; (b) M. D. Hall and T. W. Hambley, Coord. Chem. Rev., 2002, 232, 49–67 CrossRef CAS; (c) R. N. Bose and E. L. Weaver, Dalton Trans., 1997, 1797–1800 Search PubMed; (d) K. Hindmarsh, D. A. House and R. van Eldik, Inorg. Chim. Acta, 1998, 278, 32–42 Search PubMed.
- L. Chen, P. F. Lee, J. D. Ranford, J. J. Vittal and S. Y. Wong, Dalton Trans., 1999, 1209–1212 Search PubMed.
- (a) A. Casini, C. Hartinger, C. Gabbiani, E. Mini, P. J. Dyson, B. K. Keppler and L. Messori, J. Inorg. Biochem., 2008, 102, 564–575 CrossRef CAS; (b) B. Bertrand, M. R. M. Williams and M. Bochmann, Chem.–Eur. J., 2018, 24, 11840–11851 Search PubMed.
- I. V. Mironov, V. Y. Kharlamova and D. B. Kal’nyi, Russ. J. Inorg. Chem., 2024, 69, 907–913 Search PubMed.
- J. Lemke, A. Pinto, P. Niehoff, V. Vasylyeva and N. Metzler-Nolte, Dalton Trans., 2009, 7063–7070 RSC.
- I. Ott, Coord. Chem. Rev., 2009, 253, 1670–1681 Search PubMed.
- (a) C. Gabbiani, G. Mastrobuoni, F. Sorrentino, B. Dani, M. P. Rigobello, A. Bindoli, M. A. Cinellu, G. Pieraccini, L. Messori and A. Casini, MedChemComm, 2011, 2, 50–54 RSC; (b) J.-J. Jia, W.-S. Geng, Z.-Q. Wang, L. Chen and X.-S. Zeng, Toxicol. Appl. Pharmacol., 2019, 84, 453–470 Search PubMed.
- J. O. Pinho, M. Coelho, C. Pimpão, J. Konwar, A. Godinho-Santos, R. M. Noiva, S. R. Thomas, A. Casini, G. Soveral and M. M. Gaspar, Pharmaceutics, 2024, 16, 1566–1585 CrossRef CAS PubMed.
- L. Messori, F. Abbate, G. Marcon, P. Orioli, M. Fontani, E. Mini, T. Mazzei, S. Carotti, T. O'Connell and P. Zanello, J. Med. Chem., 2000, 43, 3541–3548 Search PubMed.
- T. Zou, C. T. Lum, S. S.-Y. Chui and C.-M. Che, Angew. Chem., Int. Ed. Engl., 2013, 52, 2930–2933 CrossRef CAS.
- X.-Q. Zhou, S. Abyar, I. Carbo-Bague, L. Wang, S. Türck, M. A. Siegler, U. Basu, I. Ott, R. Liu, A. P. IJzerman and S. Bonnet, CCS Chem., 2024, 6, 783–797 CrossRef CAS.
- I. V. Mironov and V. Y. Kharlamova, ChemistrySelect, 2023, 8, e202301337 CrossRef CAS.
- A. N. Wein, A. T. Stockhausen, K. I. Hardcastle, M. R. Saadein, S. B. Peng, D. Wang, D. M. Shin, Z. G. Chen and J. F. Eichler, J. Inorg. Biochem., 2011, 105, 663–668 CrossRef CAS PubMed.
- L. Skos, C. Schmidt, S. R. Thomas, M. Park, V. Geiger, D. Wenisch, R. Bonsignore, G. Del Favero, T. Mohr, A. Bileck, C. Gerner, A. Casini and S. M. Meier-Menches, Cell Rep. Phys. Sci., 2024, 5, 102072 CrossRef CAS.
- R. Kumar and C. Nevado, Angew. Chem., Int. Ed. Engl., 2017, 56, 1994–2015 Search PubMed.
- B. Salehi, Y. Berkay Yılmaz, G. Antika, T. Boyunegmez Tumer, M. Fawzi Mahomoodally, D. Lobine, M. Akram, M. Riaz, E. Capanoglu, F. Sharopov, N. Martins, W. C. Cho and J. Sharifi-Rad, Biomolecules, 2019, 9, 356–380 CrossRef CAS PubMed.
- S. W. Jordan and J. E. Cronan, J. Biol. Chem., 1997, 272, 17903–17906 CrossRef CAS PubMed.
- (a) H. Moini, L. Packer and N.-E. L. Saris, Toxicol. Appl. Pharmacol., 2002, 182, 84–90 CrossRef CAS; (b) A. Solmonson and R. J. DeBerardinis, J. Biol. Chem., 2018, 293, 7522–7530 CrossRef CAS PubMed.
- L. Rocchigiani, J. Fernandez-Cestau, I. Chambrier, P. Hrobárik and M. Bochmann, J. Am. Chem. Soc., 2018, 140, 8287–8302 CrossRef CAS PubMed.
- L. Rocchigiani, W. T. Klooster, S. J. Coles, D. L. Hughes, P. Hrobárik and M. Bochmann, Chem.–Eur. J., 2020, 26, 8267–8280 CrossRef CAS PubMed.
- N. Kuwamura, K. Hayashida, K. Tsuge, N. Yoshinari and T. Konno, Chem. Lett., 2014, 43, 1846–1848 Search PubMed.
- D. Śmiłowicz, J. C. Slootweg and N. Metzler-Nolte, Mol. Pharm., 2019, 16, 4572–4581 Search PubMed.
- M. Joost, L. Estévez, K. Miqueu, A. Amgoune and D. Bourissou, Angew. Chem., Int. Ed. Engl., 2015, 54, 5236–5240 Search PubMed.
- P. Gualco, S. Ladeira, K. Miqueu, A. Amgoune and D. Bourissou, Angew. Chem., Int. Ed. Engl., 2011, 50, 8320–8324 CrossRef CAS PubMed.
- R. E. Bachman, S. A. Bodolosky-Bettis, C. J. Pyle and M. A. Gray, J. Am. Chem. Soc., 2008, 130, 14303–14310 Search PubMed.
- M. Joost, A. Amgoune and D. Bourissou, Angew. Chem., Int. Ed. Engl., 2015, 54, 15022–15045 CrossRef CAS.
- I. V. Mironov, V. Y. Kharlamova and J. Hu, Russ. J. Inorg. Chem., 2023, 68, 287–293 CrossRef CAS.
- D. Fan, C.-T. Yang, J. D. Ranford, J. J. Vittal, P. Foo Lee and J. Russ, Inorg. Chem., 2003, 3376 CAS.
- Handbook of biochemistry and molecular biology: Physical and Chemical Data, ed. G. D. Fasman, CRC Press, Boca Raton, FL, 3rd edn, 1976, vol. 1, pp. 122–130. Search PubMed.
- A. Bast and G. R. M. M. Haenen, Biofactors, 2003, 17, 207–213 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr. A, Found. Adv., 2015, 71, 3–8 CrossRef.
- G. M. Sheldrick, Acta Crystallogr. C Struct. Chem., 2015, 71, 3–8 CrossRef.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 Search PubMed.
- A. P. Shaw, M. Tilset, R. H. Heyn and S. Jakobsen, J. Coord. Chem., 2011, 64, 38–47 CrossRef CAS.
- (a) CCDC 2424404: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2mcsld; (b) CCDC 2424406: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2mcsng.
| This journal is © The Royal Society of Chemistry 2026 |



