 Open Access Article
Open Access ArticleThermodynamic stability modulates chaperone-mediated disaggregation of α-synuclein fibrils
Celia
Fricke†
 a,
Antonin
Kunka†
a,
Antonin
Kunka†
 a,
Rasmus K.
Norrild
a,
Rasmus K.
Norrild
 a,
Shuangyan
Wang
a,
Shuangyan
Wang
 a,
Thi Lieu
Dang
a,
Thi Lieu
Dang
 b,
Jonas
Folke
b,
Jonas
Folke
 c,
Mohammad
Shahnawaz
d,
Claudio
Soto
c,
Mohammad
Shahnawaz
d,
Claudio
Soto
 d,
Susana
Aznar
d,
Susana
Aznar
 c,
Anne S.
Wentink
c,
Anne S.
Wentink
 e,
Bernd
Bukau
e,
Bernd
Bukau
 b and
Alexander K.
Buell
b and
Alexander K.
Buell
 *a
*a
aDepartment of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads, Building 227, 2800 Kgs. Lyngby, Denmark. E-mail: alebu@dtu.dk
bCenter for Molecular Biology of Heidelberg University (ZMBH), DKFZ-ZMBH Alliance, Heidelberg, Germany
cCentre for Neuroscience and Stereology, Department of Neurology, Copenhagen University Hospital, Bispebjerg and Frederiksberg Hospital, Nielsine Nielsens Vej 6B, Entrance 11B, Copenhagen, DK-2400, Denmark
dMitchell Center for Alzheimer's Disease and Related Brain Disorders, Department of Neurology, University of Texas McGovern Medical School at Houston, Houston, TX, USA
eLeiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333 CC Leiden, Netherlands
First published on 11th November 2025
Abstract
Aggregation of the intrinsically disordered protein alpha-synuclein into amyloid fibrils and their subsequent intracellular accumulation are hallmark features of several neurodegenerative disorders, including Parkinson's disease, for which no curative treatments currently exist. In this study, we investigate the relationship between fibril morphology, thermodynamic stability, and susceptibility to disaggregation by the human chaperone system comprising HSP70, DNAJB1, and Apg2. By varying assembly conditions and incubation times, we generated alpha-synuclein fibrils with diverse morphological and biochemical properties, including a broad range of thermodynamic stabilities, which we quantified using a chemical depolymerization assay. The chaperone system effectively disaggregated three of the four fibril types, with efficiencies that correlated with their thermodynamic stabilities. One fibril type resisted disaggregation despite exhibiting a comparable stability to those that were disaggregated, suggesting that additional structural features influence chaperone susceptibility. Our findings establish a quantitative link between fibril stability and chaperone-mediated disaggregation for three in vitro αSyn fibril types as well as fibrils amplified from brain extracts of PD but not MSA patients, highlighting the importance of fibril thermodynamics in biologically relevant disaggregation processes and disease pathology.
Introduction
One of the hallmark characteristics of neurodegenerative diseases such as Alzheimer's (AD) and Parkinson's (PD) is the aberrant accumulation of protein aggregates in the form of amyloid fibrils within the brain.1–4 Although the direct causal relationship between protein aggregation and disease progression remains disputed,5–7 the neurotoxic effects of amyloid fibrils, e.g. synaptic dysfunction and neuronal death, have been clearly demonstrated across various disease models.8–11 Interestingly, the main protein constituents of the extracellular and intracellular deposits in AD (amyloid-beta peptide (Aβ), Tau) and PD (α-synuclein (αSyn)) are also implicated in multiple other neurodegenerative disorders, each with distinct molecular and pathological signatures.10,12 A compelling hypothesis suggests that the distinct structures of amyloid fibrils, such as those of Tau and αSyn isolated from patients' brains, could serve as signatures linked to the underlying pathology through amyloid polymorphism.12–14 The term polymorphism refers to the ability of a single protein sequence to adopt various amyloid conformations, which differ in (i) the specific interactions within the filament core, (ii) the packing arrangement of the filament core, or (iii) the interface and organization of the protofilaments within the mature fibril.15–18Amyloid polymorphism has been observed in most amyloid-forming proteins, including Aβ, Tau, αSyn, insulin, or glucagon.18–23 This variability is considered a manifestation of the rugged amyloid energy landscape allowing multiple conformations to emerge and coexist under a given set of solution conditions, as seen for prions,24 αSyn,25,26 insulin,27 and other amyloid proteins. Polymorphism is highly sensitive to both intrinsic factors (e.g., mutations,28,29 posttranslational modifications30–32) and extrinsic factors (e.g. pH,33 ionic strength34). Time is also critical, as amyloid fibrils mature structurally over months (and potentially years).35 Despite age being the highest risk factor in most of the neurodegenerative diseases,36 most mechanistic aggregation studies focus on short timescales (i.e., minutes to days). Considerably less is known regarding the long-term (months to years) structural or morphological changes of amyloid fibrils, such as those observed from proteins like glucagon,21 amyloid beta,37,38 or αSyn.35,39,40 Similarly, recent cryo-EM studies showed time-dependent development of Tau41 and IAPP42 polymorphism during their formation and maturation, clearly demonstrating that amyloid fibril evolution on timescales beyond those of the “classical aggregation assays” (typically a few days) should be considered. It can be hypothesized that the “late stage” polymorphs persist in the brain due to their high thermodynamic stability. The stability varies significantly between fibrils formed by different amyloid peptides43 and between polymorphs,44 with reported values ranging from −65 kJ mol−1 to −14 kJ mol−1.43
The structural variability of amyloid fibrils is modulated by the cellular environment and may be linked to disease pathology either directly, through structure–toxicity relationship, or indirectly, via distinct interactions with other cellular components. Molecular chaperones are essential in this context, serving as protein quality control and maintaining cellular homeostasis.45–47 Fundamental aspects of chaperone action include assisting protein folding, preventing protein aggregation and targeting proteins to degradation machineries such as the ubiquitin/proteasome system or the autophagy system.48–51 A specific chaperone system consisting of Heat Shock Protein 70 (HSP70) and co-chaperones from the J-domain protein (JDP) and Nucleotide Exchange Factor (NEF) families can disaggregate amyloid fibrils formed by αSyn52–55 Tau,56 and exon1 of the huntingtin protein.57 The mechanism is ATP-dependent and involves generating an entropic pulling force.52,58–60 HSP70 and JDP DNAJB1 mainly interact with the flexible regions in the N- and C-terminus of αSyn fibrils and not with the fibril core.52 Distinct polymorphs of αSyn fibrils are disaggregated to varying degrees, which has been attributed to an interplay between the chaperones' affinities for the fibrils and variations in the polymorph's stabilities.61 In contrast to the chaperone binding, the stabilities have only been inferred indirectly from the fibrils' resistance to cold denaturation, and their precise quantification is still missing. Furthermore, how amyloid thermodynamic stability, fibril polymorphism, and their temporal changes influence the efficiency of disaggregation by chaperones remains unclear.61
Here, we investigate the link between fibrils' morphology, thermodynamic stability, and their susceptibility to disaggregation by the human tri-chaperone system. We prepare αSyn fibrils at four different solution conditions over a timespan ranging from three days to three months and characterize them using a series of biochemical and biophysical assays. The resulting set of 24 fibril preparations differ in structural and morphological features and display a wide range of thermodynamic stabilities. Importantly, we demonstrate that their stability is negatively correlated with the disaggregation efficiency of the tri-chaperone system HSP70, DNAJB1, and Apg2, for three out of four fibril types studied here, and the fourth type is never depolymerized. The correlations are sensitive to the specific fibril types, indicating that apart from the thermodynamic stability of the fibrils, their structural or morphological features influence the action of the chaperones. These conclusions are further supported by analysis of more physiologically relevant fibrils formed by amplification of seeds from brain extracts of MSA and PD patients. Altogether, our results provide important insights into the interplay between fibril polymorphism, stability, and chaperone disaggregation efficiency.
Results
Selection of assembly conditions and experimental design
To generate distinct αSyn polymorphs, we selected four in vitro experimental solution conditions that have been used previously to obtain well-defined sets of fibril morphologies termed Fibrils (Fm), Ribbons (Ri), Fibrils-65 (F65) and Fibrils-91 (F91) (Table 1).34,62–64 Two polymorphic structures, 2a (pdb id 6ssx) and 2b (pdb id 6sst), have been resolved in the Fm samples using cryo-EM,65 whereas only a protein secondary structure assignment is available for Ri and F91 based on solid-state NMR experiments.64,66 The structure of F65 is unknown but their twisted morphology, and proteinase K digestion fingerprint resemble those of the Fm.62 To specifically investigate the effect of incubation time, we prepared samples at four sets of solution conditions while keeping protein concentration, temperature, shaking speed, and sample volumes constant to minimize variation stemming from these sources (Fig. 1). We then collected and characterized fibrils at six different time points ranging from 3 days to 3 months, to assess how incubation time influences fibril stability and susceptibility to disaggregation by chaperones. Altogether, we carried out three independent repeats of the experiment using two distinct monomer protein batches, resulting in a total of 72 samples (24 samples per experiment). This experimental design yielded a robust dataset for studying the relationship between fibril stability and chaperone-mediated disaggregation within the context of a single protein sequence.| Condition | Buffer composition | Incubation time (days) and fibril sample availability per repeat | |||||
|---|---|---|---|---|---|---|---|
| 3 days | 6 days | 14 days | 28 days | 56 days | 84 days | ||
| Fm | 50 mM Tris-HCl, 150 mM KCl, 1 mM EDTA, 0.05% NaN3, pH 7.5 | ✗✗✗ | ✗✗✗ | ✓✓✓ | ✓✓✓ | ✓✓✓ | ✓✗✓ |
| Ri | 5 mM Tris-HCl, 1 mM EDTA, 0.05% NaN3, pH 7.5 | ✗✗✗ | ✗✗✗ | ✗✓✓ | ✓✓✓ | ✓✓✗ | ✓✓✓ |
| F65 | 20 mM MES, 150 mM NaCl, 1 mM EDTA, 0.05% NaN3, pH 6.5 | ✓✗✗ | ✓✓✓ | ✓✓✓ | ✓✓✓ | ✓✓✓ | ✓✓✓ |
| F91 | 20 mM KPO4, 1 mM EDTA, 0.05% NaN3, pH 9.1 | ✗✗✗ | ✗✗✗ | ✗✗✗ | ✓✓✗ | ✗✓✗ | ✗✓✗ |
 | ||
| Fig. 1 Overview of the experimental design. Left panel: αSyn monomer was brought to four different sets of solution conditions – physiological (Fm, blue), low salt (Ri, orange), mildly acidic (‘low’) pH (F65, green), and mildly basic (‘high’) pH (F91, violet). Middle panel: sample aliquots (0.5 mL, 100 µM for each timepoint and condition) were incubated at 37 °C and 600 rpm for 3, 6, 14, 28, 56, and 84 days. All samples were analyzed simultaneously by the indicated assays described in detail in the materials and methods section. Right panel: schematic overview of the relation of fibril stability and susceptibility to the chaperone system Hsp70, DNAJB1 and Apg2. The energy diagram illustrates that highly disordered monomeric αSyn with high energy assembles into the thermodynamically more favorable fibrillar state. Different fibrils can have different thermodynamic stability, indicated by the purple and green states, and can be depolymerized by tri-chaperone system to varying degrees. The stability and chaperone disaggregation of the fibrils from each sample were analyzed using established methods.44,52,56 | ||
Analysis of soluble fraction and fibril characterization
First, we removed the supernatant from all samples by centrifugation and quantified the residual monomer concentration by UV-absorbance (SI Fig. 1a). We obtained comparable absorbance values with a benchtop centrifuge (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900×g) and ultracentrifuge (220
900×g) and ultracentrifuge (220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g), confirming that the insoluble fraction was successfully pelleted (SI Fig. 1b). The Fm and F65 samples aggregated rapidly within two weeks of incubation, whereas the aggregation of Ri and F91 took approximately four weeks to reach a plateau. The apparent monomer concentration converged to approximately 10–20 µM for all conditions except F91, for which it was significantly higher (60 µM). To rule out the possibility of a pseudo-equilibrium state caused by higher-order assembly of fibrils (observed in most of the tubes) which might impair their further growth, we sonicated aliquots from the samples after 84 days of incubation to break down possible fibril clumps and generate more seeding-competent fibril ends and then extended the incubation period by five days. The monomer concentration in the supernatants of the centrifuged samples after this period did not change significantly in all conditions (cross marks in SI Fig. 1a). Interestingly, we did not observe any significant dissociation of the fibrils back into monomers (re-equilibration) when we removed the supernatant and resuspended fibrils in new buffer (SI Fig. 2), indicating that the measured apparent monomer concentration during fibril preparation does not reflect the true equilibrium. To investigate further, we supplemented the isolated supernatants with new wild-type seeds (prepared from an independent fibril batch in condition Fm). We observed that the long incubation times lead to reduced or completely abolished elongation efficiency of the soluble fraction (SI Fig. 3). SDS-PAGE together with mass spectroscopy (MS) analysis revealed degradation of αSyn in the aged (>1 month) samples, but no substantial chemical modifications (SI Fig. 4–10 and SI File 1). The degree of degradation varied between experimental repeats (1st < 2nd < 3rd) and was most prominent in the F91 samples (SI Fig. 1, 4 and 9). Taken together, these findings suggest that the final fibril yield is not a direct measure of αSyn solubility; instead, it is influenced by degradation of soluble species which limits their incorporation into fibrils. Next, we resuspended the centrifuged fibrils from each sample in their corresponding buffers, and characterized their secondary structures, amyloid dye sensitivity, seeding properties, morphology, and resistance to proteolysis using circular dichroism (CD) spectroscopy (SI Fig. 11 and 12), ThT fluorescence (SI Fig. 13 and 14), atomic force microscopy (AFM) (SI Fig. 15 and 16), and proteinase K digestion (SI Fig. 17), respectively. Based on our analyses, we conclude that fibrils formed under the four selected conditions exhibit distinct structural and morphological characteristics, albeit not identical in all cases to those reported previously for the same conditions.34,62–64 These characteristics evolve with incubation time, as described for αSyn35,40 and other pathological amyloids such as Tau41 or IAPP,42 with the most pronounced changes observed at neutral (Fm) or mildly acidic (F65) pH under physiological salt concentrations. These conditions promote αSyn polymorphism more strongly than the low-salt (Ri) or high-pH (F91) conditions, which each favor the formation of distinct yet more internally consistent fibril structures. A detailed description of the morphological and structural characterization of the fibrils is provided in the SI.
000×g), confirming that the insoluble fraction was successfully pelleted (SI Fig. 1b). The Fm and F65 samples aggregated rapidly within two weeks of incubation, whereas the aggregation of Ri and F91 took approximately four weeks to reach a plateau. The apparent monomer concentration converged to approximately 10–20 µM for all conditions except F91, for which it was significantly higher (60 µM). To rule out the possibility of a pseudo-equilibrium state caused by higher-order assembly of fibrils (observed in most of the tubes) which might impair their further growth, we sonicated aliquots from the samples after 84 days of incubation to break down possible fibril clumps and generate more seeding-competent fibril ends and then extended the incubation period by five days. The monomer concentration in the supernatants of the centrifuged samples after this period did not change significantly in all conditions (cross marks in SI Fig. 1a). Interestingly, we did not observe any significant dissociation of the fibrils back into monomers (re-equilibration) when we removed the supernatant and resuspended fibrils in new buffer (SI Fig. 2), indicating that the measured apparent monomer concentration during fibril preparation does not reflect the true equilibrium. To investigate further, we supplemented the isolated supernatants with new wild-type seeds (prepared from an independent fibril batch in condition Fm). We observed that the long incubation times lead to reduced or completely abolished elongation efficiency of the soluble fraction (SI Fig. 3). SDS-PAGE together with mass spectroscopy (MS) analysis revealed degradation of αSyn in the aged (>1 month) samples, but no substantial chemical modifications (SI Fig. 4–10 and SI File 1). The degree of degradation varied between experimental repeats (1st < 2nd < 3rd) and was most prominent in the F91 samples (SI Fig. 1, 4 and 9). Taken together, these findings suggest that the final fibril yield is not a direct measure of αSyn solubility; instead, it is influenced by degradation of soluble species which limits their incorporation into fibrils. Next, we resuspended the centrifuged fibrils from each sample in their corresponding buffers, and characterized their secondary structures, amyloid dye sensitivity, seeding properties, morphology, and resistance to proteolysis using circular dichroism (CD) spectroscopy (SI Fig. 11 and 12), ThT fluorescence (SI Fig. 13 and 14), atomic force microscopy (AFM) (SI Fig. 15 and 16), and proteinase K digestion (SI Fig. 17), respectively. Based on our analyses, we conclude that fibrils formed under the four selected conditions exhibit distinct structural and morphological characteristics, albeit not identical in all cases to those reported previously for the same conditions.34,62–64 These characteristics evolve with incubation time, as described for αSyn35,40 and other pathological amyloids such as Tau41 or IAPP,42 with the most pronounced changes observed at neutral (Fm) or mildly acidic (F65) pH under physiological salt concentrations. These conditions promote αSyn polymorphism more strongly than the low-salt (Ri) or high-pH (F91) conditions, which each favor the formation of distinct yet more internally consistent fibril structures. A detailed description of the morphological and structural characterization of the fibrils is provided in the SI.
Thermodynamic stability varies across distinct α-Synuclein fibril polymorphs
To assess the thermodynamic stability of αSyn fibrils, we used urea depolymerization.44,67 The urea depolymerization was performed in the buffer used for chaperone disaggregation (50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT) to allow its direct comparison to the thermodynamic stability. The fibrils were centrifuged, the supernatant removed, and the fibril pellet resuspended and sonicated to achieve consistent concentration (within <10% error) and homogeneity for all samples. Using CD spectroscopy and proteinase K, we confirmed that resuspension in the assay buffer for chaperone disaggregation does not alter the secondary structure of the fibrils (SI Fig. 18 and 19). The fraction of soluble monomer at increasing concentrations of urea was determined using flow-induced dispersion analysis (FIDA) as previously described44 and fitted using the isodesmic polymerization model.68 We employed Hamiltonian Monte Carlo (HMC) sampling of solutions and Bayesian analysis to better capture the relationship between the stability (ΔG) and the sensitivity towards the denaturant (i.e., the m-value) which were found to be correlated in conventional fitting (SI Fig. 20–23 and SI Tables 1–3).69 First, we used HMC sampling of solutions to evaluate ΔG in an unbiased manner (SI Fig. 20–23a). The HMC sampling clearly revealed the correlation between ΔG and the m-values in most cases. To reduce this dependency, we carried out the analysis with a moderate prior on the m-value (mean = 3.5 kJ mol−1 M−1, σ = 2, SI Fig. 20–23b). The prior was chosen based on the estimated changes of solvent accessible area between monomers in the amyloid and free state assuming it relates to the urea m-value analogously to what has been observed for unfolding of globular proteins (see Materials and Methods section for details of the calculation).70,71 The Bayesian analysis enabled more confident estimation of individual ΔG values (Fig. 2a, b and SI Fig. 20–23b). Generally, we observed that ΔG decreased with incubation time for most datasets, albeit the negative slopes were statistically significant only for two repeats of F65 (marked by asterisk in Fig. 2c and SI Table 5). Regardless of the ΔG(t) relationship, the thermodynamic stability of individual fibril preparations varied considerably from −26 to −43 kJ mol−1, making it a well-suited dataset for studying the relationship between αSyn fibril stability and chaperone-mediated disaggregation. | ||
| Fig. 2 Thermodynamic stability of fibrils formed in conditions Fm, Ri, F65 and F91 and measured in 50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT. (a) Fitting of the isodesmic model to depolymerization curves of amyloid fibrils formed under condition F65 at various time points (color-coded as indicated in the legend) using Bayesian analysis. The prior distribution for the m-values was defined by a mean of 3.5 and a standard deviation of 2. Opaque lines represent 100 random samples from a Hamiltonian Monte Carlo (HMC) analysis, drawn from at least 2000 total samples. The area under each curve (AUC) corresponds to the monomer concentration measured by FIDA.44 (b) Joint probability distributions of ΔG and m-values obtained from the fitting in (a), highlighting the correlation between these two fitting parameters. (c) Thermodynamic stability (ΔG) of fibrils formed at different incubation times, determined from urea-induced depolymerization experiments. Depolymerization curves were fitted to the isodesmic model using Bayesian analysis as demonstrated in (a and b). Error bars represent the uncertainty (mean ± standard deviation) derived from the HMC analysis (n = 2000 samples). Repeats with significant (p < 0.05) negative slopes are marked with a red asterisk. Parameter values (ΔG and m-values) are listed in SI Tables 1–3. | ||
Thermodynamic stability is an important, but not the sole determinant of fibril disaggregation by chaperones
To investigate the importance of fibril stability on the disaggregation by chaperones, we subjected the fibrils formed in the four different conditions to the chaperone system consisting of HSP70, DNAJB1 and Apg2 in the same buffer system (50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM ATP, 2 mM DTT), and used ThT to monitor their disaggregation (Fig. 3 and SI 24).52,56 The fibrils were centrifuged, the supernatant removed, and the fibril pellet resuspended and sonicated to achieve consistent concentration and homogeneity across all conditions and time points. All fibril types were disaggregated with varying efficiencies except for Ri fibrils which were found to be resistant to chaperone action in all but one sample (Fig. 3, SI Fig. 24, 25 and SI Table 6). We further confirmed the minimal Ri disaggregation by western blot analysis of the supernatant fraction upon 18 h incubation with chaperones with and without ATP (SI Fig. 26). In our previous report, we attributed the lack of Ri disaggregation to fibril stability inferred from resistance of fibrils to cold denaturation.61 Here, we observe no appreciable dissociation of any of the investigated fibril types upon their three-day incubation at 4 °C (SI Fig. 2). This lack of detectable cold-depolymerisation is likely due to the high absolute stability72 and highlights the value of chemical depolymerisation that we employ as our main quantification strategy for thermodynamic stability in the present study. However, our quantitative analysis clearly shows that thermodynamic stability is not the principal determinant of Ri fibrils' resistance to chaperones since other fibrils with a similar stability are disaggregated (Fig. 5).Combining all available data together, we find a correlation between fibril stability and their degree of disaggregation for the other three fibril types, i.e., Fm, F65 and F91 (r of 0.66, 0.74 and 0.92 and p-value of 0.03, 0.001 and 0.08, respectively, Fig. 5). The disaggregation efficiency exhibited a decreasing trend with fibril maturation time for all three repeats of F65 and one repeat of F91, and no apparent dependency for Fm (Fig. 3b). The F65 samples were the most consistent in terms of changes in fibril stability and chaperone disaggregation (Fig. 2 and 3), as well as structural properties across the three biological repeats (SI Fig. 11, 12, and 15). Arguably, the difference in stability and disaggregation between early (3 days) and late (2–3 months) fibrils, along with the negative correlation observed, can thus be directly linked to fibril maturation in the F65 condition. Mature fibrils were approximately 10 kJ mol−1 more stable and 50% less susceptible to disaggregation than the early fibrils. CD and AFM analyses revealed an increase in beta-sheet content, fibril height, and apparent pitch length over time (SI Fig. 11, 12 and 15), suggesting that the optimized interaction network between residues and/or improved protofilament packing may account for these differences. Profiles obtained by limited proteolysis of 14- and 84 days old F65 fibrils revealed only minor differences at early time-points, supporting the conclusion that the increased stability and resistance to disaggregation is more likely due to changes in the structured core rather than the organization and accessibility of the disordered fuzzy coat (SI Fig. 17). A similar trend was observed for F91 fibrils, which became more stable and resistant to disaggregation over time (Fig. 2c and 3b). The relatively high increase in stability (∼5 kJ mol−1) was accompanied by only a moderate change in chaperone disaggregation (ca 15%). In contrast to F65, the disordered regions of 84 days old F91 fibrils were less susceptible to proteolysis compared to 14 days old ones, suggesting that fuzzy coat accessibility affects the chaperone disaggregation efficiency. However, assessing variability across repeats was difficult for F91 due to low fibril yield resulting from the high solubility of αSyn and its susceptibility to degradation in this condition (Table 1).
In contrast, the Fm samples exhibit considerable variability between repeats in most of our assays, likely due to the polymorph heterogeneity under physiological conditions.73 Although the effect of incubation time on fibril stability and chaperone disaggregation is complicated by stochastic factors that influence the emergence of specific fibril polymorphs in each sample, the stability of the resulting fibrils and their susceptibility to chaperone disaggregation show good correlation, regardless of the polymorphs formed (Fig. 5). Both properties varied considerably between individual samples, with their correlation falling within a similar range and exhibiting sensitivity comparable to that of F65. This suggests a potential overlap of the polymorphic landscape of αSyn at the two conditions leading to fibrils with similar structural and morphological features and, consequently, sensitivities to chaperone degradation. AFM analysis of Fm samples from the first repeat reveals a heterogeneous mixture of twisted and non-twisted fibrils, with some subpopulations exhibiting similar height and apparent pitch length to F65 fibrils (SI Fig. 15), supporting a certain degree of similarity between Fm and F65.
To extend our observations to more physiologically relevant fibril polymorphs, we analyzed the stability and chaperone disaggregation of fibrils amplified from brains of Parkinson's disease (PD) and Multiple System Atrophy (MSA) patients using a αSyn seeding amplification assay (αSyn-SAA) (Fig. 4 and SI Fig. 27). Although it has been proposed that the structure of the original fibrils may not necessarily be fully propagated in SAAs in all cases,74 the resulting amplified products clearly show distinct polymorph populations.75 The PD derived fibrils were less stable (−27.53 ± 1.63 kJ mol−1) compared to the MSA-derived ones (−32.0 ± 1.63 kJ mol−1) (Fig. 4a and SI Table 4). The PD-derived fibrils were disaggregated with moderate efficiency (45%) whereas the more stable MSA-derived ones were not disaggregated by the chaperone system (Fig. 4b). The thermodynamic stability and chaperone disaggregation of fibrils amplified from patients' brains show similar correlation to polymorphs Fm, F65 and F91 prepared in vitro, i.e., less disaggregation for more stable fibrils. Simultaneously, stability alone cannot explain the lack of disaggregation as polymorphs more stable than MSA-derived fibrils get disaggregated (Fig. 5). This further highlights that other factors, such as fibril surface exposure or chaperone binding mode may play a role in determining disaggregation efficiency.
Altogether, we conclude that fibril stability is an important, but not the sole factor influencing the fibrils' susceptibility to chaperone disaggregation. Our study provides a new quantitative link between fibril stability and chaperone-mediated disaggregation for three out of four in vitro fibril types and fibrils amplified from brain extracts of PD, but not MSA patients, further highlighting the role of fibril morphology in determining the efficacy of chaperone action.
Discussion and conclusions
Synucleinopathies are characterized by the aggregation and accumulation of αSyn fibrils which progress over several years. The fibril deposits are highly heterogenous and change over time, undergoing structural and morphological changes as well as being subjected to post-translational modifications and truncations, which in turn modulate how efficiently they are cleared by the homeostasis control systems of the cells.76The efficiency of the chaperone system HSP70/DNAJB1/Apg2 towards disaggregation of αSyn fibrils is sensitive to amyloid polymorphism, suggesting important roles of structural features and fibril stability.61
Here, we explored the relationship between changes in amyloid fibril polymorphism, thermodynamic stability, and the susceptibility of αSyn fibrils to disaggregation by the tri-chaperone system occurring during their long-term incubation in vitro. The variance of the results across the three biological repeats of the time-dependent changes during fibril formation over extended periods allowed us to gain important insights into the polymorphic landscape of αSyn. We found that the decrease in soluble protein was consistent across the repeats (SI Fig. 1a) whereas the heterogeneity of the fibrils was highly sensitive to extrinsic conditions. Specifically, the high consistency of F65 samples suggests that the stochastic effects influencing nucleation and growth are less pronounced, likely due to a more funneled nature of the energy landscape at low pH (i.e., F65) compared to the physiological conditions (i.e., Fm).40,73 Consequently, we can infer the time-dependent morphological changes and their effects on stability and chaperone disaggregation for F65 samples with high confidence. In contrast, fibrils formed under other conditions (i.e., Fm, Ri) show greater variability, making it more challenging to discern clear time-dependent trends without more detailed structural analysis of the individual samples. Our morphological analysis conducted using AFM on samples from the first repeat revealed significant changes in the apparent pitch length of fibrils formed under physiological salt concentrations (Fm) and low pH conditions (F65), along with variations in height across all four conditions over time. This observation, together with altered ThT sensitivity, changes in secondary structure, and variations in seeding efficiency (SI Fig. 13 and 14) suggest significant shifts in the distribution of polymorph populations between samples incubated at different timescales, consistent with earlier reports on αSyn and other amyloids.35 These may include differences in the exposure of fibril core, packing, and/or dynamics of the flexible termini (i.e., fuzzy coat). The fuzzy coat is especially important as it contains chaperone binding sites.52,77,78 We were unable to detect any striking differences in the SDS-PAGE profiles of proteinase K digestion (SI Fig. 17) that would help explain the varying degrees of fibril disaggregation. Interestingly, the fuzzy coat of polymorph Ri that resists depolymerization by chaperones is quickly degraded by proteinase K (SI Fig. 17 and 18). Previous studies showed that the N-terminus of Ri fibrils is more structured, less solvent exposed and therefore protected, whilst the flexible C-terminus (∼95–140) gets degraded by the protease.61,79 We therefore hypothesize that the degree of disorder and accessibility of chaperone binding sites within the N-termini in the fuzzy coat of the αSyn fibrils could be important factors of chaperone disaggregation efficiency. Chaperone disaggregation experiments with fibrils from N-terminally truncated αSyn variants, together with higher resolution analysis of more uniform population of fibrils will be needed to dissect the effect of the fuzzy coat on chaperone disaggregation efficiency in future studies. Different patterns of fibril lateral association or higher order assembly (e.g., fibril clumping) between the polymorphs might also affect the availability of the fibril surface or fibril ends.55 Extending the experimental characterization by classification of the distinct polymorph populations using cryo-electron microscopy in the future will allow to model the development of polymorphism over time and will provide a more detailed link between fibril structure, stability, and susceptibility to chaperone disaggregation.
We find that under some conditions, notably at slightly basic pH (9.1) αSyn undergoes degradation at longer incubation times (SI Fig. 4–10). This includes degradation of free monomers, as well as truncation of the already formed fibrils. Given that the chaperones HSP70 and DNAJB1 are thought to interact primarily with the C- and N-terminal disordered regions of the fibrils, one might argue that the truncations are a main determinant for the observed change in degree of depolymerisation. However, F91 fibrils are easily depolymerised by the chaperones despite their tendency to undergo truncation. Furthermore, while truncation could be a confounding factor in some cases, it does occur in vivo to a significant extent.80–83 Therefore, our observations that both thermodynamic stability and depolymerisation are clearly correlated with each other does not exclude that both of these properties are also correlated to the degree of truncation. Whether there is a causal relationship or merely a correlation between all these effects will need to be examined in future studies.
Given that most tyrosine residues are in the C-terminus, even a modest degree of C-terminal truncation could lead to a misrepresentation of the absorbance measurements of the soluble monomer fraction at equilibrium. Moreover, we observe that the soluble fraction ceases to be elongation competent after ca 1 month of incubation (SI Fig. 2). This suggests that the degradation products are unable to incorporate into fibrils and/or interfere with incorporation of the residual intact monomer (SI Fig. 4–8). In contrast, the pelleted fibrils maintain seeding capacity and can be elongated by fresh monomer (SI Fig. 13 and 14), which is an important prerequisite of the chemical depolymerization assay. The Bayesian analysis of the chemical depolymerization allowed us to explore the relationship between the ΔG and the m-values in detail. Interestingly, the average m-values obtained from fits of Ri and F91 depolymerization were close to the prior (ca 2–3.5 kJ mol−1 M−1, respectively) compared to the Fm and F65 which were significantly higher (ca 6 and 8 kJ mol−1 M−1, respectively). Notably, the m-values correlated weakly (R2 = 0.3) with AFM-derived fibril heights, suggesting that the m-values obtained from the isodesmic model may indeed reflect structural properties of the fibrils. The differences observed here may relate to distinct solvent exposure of the flexible N- and C-termini between the polymorphs, which were not accounted for when calculating the prior value. The fibril stability ranged between ca −26 and −43 kJ mol−1, consistent with previous studies,44,69,84 providing a suitable dataset for investigating its contribution to fibril disaggregation within the context of a single protein sequence.
The results presented in this work corroborate our previous observations regarding the relationship between fibril stability and disaggregation of different αSyn polymorphs by the tri-chaperone system HSP70/DNAJB1/Apg2.61 We have found a clear correlation between ΔG and the degree of disaggregation for F65, Fm and F91, and no disaggregation of fibrils formed in the absence of salt (Ri). Our previous studies demonstrated that the lack of Ri fibril disaggregation cannot be explained by a weak chaperone binding and we attributed this observation to their high stability inferred from cold denaturation.61 Here, we extended our analysis to show that none of the fibril samples (except one) formed under these conditions undergo disaggregation, despite having a similar range of thermodynamic stability as the other fibrils that do get disaggregated. This finding clearly shows that other factors beyond fibril stability must be at play.
Our conclusions extend further to physiologically relevant fibrils obtained by amplification of seeds from patients' brain extracts, where we observed that MSA-derived fibrils are more stable and resistant to disaggregation, whereas PD-derived fibrils are less stable and get disaggregated by chaperones. The lack of MSA-amplified fibril disaggregation cannot be solely explained by their stability, further corroborating that other factors must play role. These may include dynamics or altered bound conformation of the chaperones resulting in their recruitment to the fibrils in an inactive state. Two of the clear distinguishing morphological features of Ri fibrils compared to fibrils that get disaggregated (i.e., Fm and F65) are the lack of a helical twist and high degree of structure in the N-terminus.85 This can potentially affect the spatial organization of the disordered regions along the fibrils and influence the important features of the fuzzy coat such as charge distribution. The higher-order assembly of fibrils by their lateral association, observed for both Ri and MSA-amplified fibrils that resist disaggregation, indicates that surface properties may play a critical role in determining their interactions with the chaperone system.
Importantly, our results demonstrate that the human tri-chaperone system can indeed disaggregate fibrils as stable as –40 kJ mol−1 in certain cases. This value is significantly less compared to the free energy of ATP hydrolysis at the beginning of our depolymerization assays (ΔG = −68.5 kJ mol−1, assuming 1% of initially hydrolyzed ATP, Fig. 6).86 However, the value drops to −40.3 kJ mol−1 when 90% of ATP is consumed and would recover only marginally (−46.3 kJ mol−1) upon addition of fresh 2 mM ATP to the reaction. This suggests that the extent of ATP depletion, along with the accumulation of ADP and phosphate may set a stability threshold for fibrils that can still be disaggregated by the chaperones.
 | ||
| Fig. 6 Change in the free energy of ATP as a function of its depletion. The Gibbs free energy of ATP hydrolysis to ADP and phosphate (P) (ΔG, black line) was calculated for conditions of the disaggregation assay (150 mM ionic strength, pH 7.5, 5 mM Mg2+) using the standard values from.86 The range of free energy corresponding to the fibril stabilities measured here are highlighted in red and green. | ||
Our findings hold significant implications for amyloid fibrils formed by proteins involved in other neurodegenerative diseases, such as Aβ, Tau, or IAPP. In the brain, amyloid fibrils persist over many years suggesting a high thermodynamic stability, which might lead to their resistance against disaggregation by the natural chaperone systems. Systematic studies of fibril stability and chaperone-mediated disaggregation, like the approach described here, will help to generalize our conclusions to amyloids formed from diverse protein sequences. Understanding the factors that determine the disaggregation propensity is a crucial step in understanding the molecular mechanisms in Parkinson's and other neurodegenerative diseases. Importantly, while chaperone-mediated disaggregation may help reduce amyloid load, it can also produce potentially toxic seeding competent smaller species.61 Further mechanistic studies are therefore needed to ensure that the therapeutic strategies for reducing amyloid load by fine tuning or boosting of the chaperone system (e.g., by optimization of nucleotide recycling) or increasing the susceptibility of amyloid fibrils to the chaperone system will be successful.
Methods
Protein expression and purification
The chaperones DNAJB1 and Apg2 were expressed and purified as described before.52The human constitutive, cytosolic isoform of Hsp70, Hsc70 (HSPA8) was produced as follows:
Chemo-competent E. coli BL21 (DE3) (New England Biolabs) cells were transformed by pCA528_SUMO_Hsc70 using heat shock. The expression of 6xHis-SUMO-Hsc70 was induced by IPTG (1 mM final concentration) and carried out at 20 °C for approximately 16 h. Cells were harvested by centrifugation (6000×g, 4 °C, 15 min), resuspended in working buffer (WB, 50 mM HEPES, pH 7.4, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 10 mM imidazole, 1 mM DTT) containing 1 mM PMSF and commercial DNase (Benzonase Nuclease, Sigma Aldrich), and lysed by sonication (Qsonica Q500, Newtown, CT, USA). The lysed solution was centrifuged at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 45 min at 4 °C. The supernatant was added to Ni-NTA beads (Thermo Scientific, Waltham, MS, USA) equilibrated in WB. The solution was incubated with the beads for one hour at 4 °C. Subsequently, beads were washed with WB containing 20 mM imidazole and bound proteins eluted by WB with 250 mM imidazole. Fractions containing 6xHis-SUMO-Hsp70 were cleaved by a SUMO protease (SAE0067, Merck KGaA, Darmstadt, Germany) during ca 16 h dialysis into 25 mM HEPES, pH 8.0, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 2 mM DTT at 4 °C. On the next day, the solution was added to NiNTA beads and unbound protein including HSP70 was eluted by WB containing 20 mM imidazole. Fractions containing HSP70 were loaded onto a HiTrap Q HP, 5 mL column pre-equilibrated in 50 mM HEPES, pH 8.0, 10 mM KCl, 5 mM MgCl2, 1 mM DTT, Proteins were eluted by a 0–70% gradient of 1 M KCl. Fractions containing HSP70 were pooled, concentrated and loaded onto a Superdex 200 10/300 increase GL (Cytiva) pre-equilibrated in 50 mM HEPES, pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM DTT. Fractions containing highly pure HSP70 were flash frozen and stored at −80 °C.
000 rpm for 45 min at 4 °C. The supernatant was added to Ni-NTA beads (Thermo Scientific, Waltham, MS, USA) equilibrated in WB. The solution was incubated with the beads for one hour at 4 °C. Subsequently, beads were washed with WB containing 20 mM imidazole and bound proteins eluted by WB with 250 mM imidazole. Fractions containing 6xHis-SUMO-Hsp70 were cleaved by a SUMO protease (SAE0067, Merck KGaA, Darmstadt, Germany) during ca 16 h dialysis into 25 mM HEPES, pH 8.0, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 2 mM DTT at 4 °C. On the next day, the solution was added to NiNTA beads and unbound protein including HSP70 was eluted by WB containing 20 mM imidazole. Fractions containing HSP70 were loaded onto a HiTrap Q HP, 5 mL column pre-equilibrated in 50 mM HEPES, pH 8.0, 10 mM KCl, 5 mM MgCl2, 1 mM DTT, Proteins were eluted by a 0–70% gradient of 1 M KCl. Fractions containing HSP70 were pooled, concentrated and loaded onto a Superdex 200 10/300 increase GL (Cytiva) pre-equilibrated in 50 mM HEPES, pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM DTT. Fractions containing highly pure HSP70 were flash frozen and stored at −80 °C.
αSyn expression and purification were carried out from pT7-7 αSyn WT plasmid (RRID:Addgene_36046) as described previously.44 In short, harvested cells were lysed using sonication (Qsonica Q500, Newtown, CT, USA, 10 s on time, 30 s off time, 12 rounds with 40% amplitude), and cell-free lysate further processed by boiling for 20 minutes. The resulting solution was precipitated by saturated (NH4)2SO4 (4 mL per 1 mL of supernatant, 15 min at 4 °C) and centrifuged (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 20 min at 4 °C). The resulting pellet was resuspended in 7 mL of 25 mM tris-HCl pH 7.7 with 1 mM DTT and dialyzed twice into the same buffer. Monomeric αSyn was purified by anion exchange chromatography (AEC) (HiTrap Q Hp 5 mL, GE Healthcare) using linear gradient of NaCl, followed by size exclusion chromatography (SEC) (HiLoad 16/600 Superdex 200 pg. column, Cytiva). The monomeric fraction of αSyn eluted in 10 mM of NaP buffer pH 7.4 was collected, flash-frozen in liquid nitrogen and stored at −80 °C.
000×g, 20 min at 4 °C). The resulting pellet was resuspended in 7 mL of 25 mM tris-HCl pH 7.7 with 1 mM DTT and dialyzed twice into the same buffer. Monomeric αSyn was purified by anion exchange chromatography (AEC) (HiTrap Q Hp 5 mL, GE Healthcare) using linear gradient of NaCl, followed by size exclusion chromatography (SEC) (HiLoad 16/600 Superdex 200 pg. column, Cytiva). The monomeric fraction of αSyn eluted in 10 mM of NaP buffer pH 7.4 was collected, flash-frozen in liquid nitrogen and stored at −80 °C.
Fibril assembly
Fibrils were prepared in four different conditions provided in Table 1. Purified αSyn monomer was buffer exchanged into the desired assembly buffer using 3 kDa cut-off spin column filter (Amicon® Ultra Centrifugal Filter, Merck). The concentration was adjusted to 100 µM and the sample in each solution condition split into 12 aliquots of 0.5 mL which were flash-frozen in liquid nitrogen and stored at −80 °C. Two aliquots from each condition were thawed and placed into the incubator (Eppendorf ThermoMixer C, Germany) at 37 °C at 600 rpm at six different time points such that their incubation periods (3, 6, 14, 28, 54, and 84 days) ended simultaneously. After 84 days, one set of aliquots per timepoint per condition (i.e., n = 24) was used directly in the experiments described further below whilst the other was flash frozen and stored at −80 °C and used as a 2nd repeat. The third repeat was carried out from an independent monomer batch analogously to the first repeat (i.e., samples measured directly without freeze-thawing).Analysis of residual monomer concentration
A 50 µL aliquot was taken from each sample and centrifuged (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 90 min, 25 °C). The resulting supernatant was carefully transferred to a clean tube and analyzed using SDS-PAGE, flow-induced dispersion analysis (FIDA), and UV-absorbance. The latter was carried out on NanoDrop instrument (ThermoFisher, USA) using the extinction coefficient of αSyn (ε280 = 5960 cm−1 M−1) calculated from the sequence using Expasy webserver.
000×g, 90 min, 25 °C). The resulting supernatant was carefully transferred to a clean tube and analyzed using SDS-PAGE, flow-induced dispersion analysis (FIDA), and UV-absorbance. The latter was carried out on NanoDrop instrument (ThermoFisher, USA) using the extinction coefficient of αSyn (ε280 = 5960 cm−1 M−1) calculated from the sequence using Expasy webserver.
For the third repeat, a 50 µL aliquot of each sample was additionally centrifuged using ultracentrifuge (Beckman Coulter-Optima XE-90). Samples were loaded to 0.2 mL, open-top thickwall polycarbonate tubes (Beckman), placed in 42.2 Ti Fixed-Angle Titanium Rotor, and centrifuged at 214![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 361×g for 2 hours at 5 °C. The supernatant was carefully removed and analyzed by UV-absorbance and SDS-PAGE as described above and below, respectively.
361×g for 2 hours at 5 °C. The supernatant was carefully removed and analyzed by UV-absorbance and SDS-PAGE as described above and below, respectively.
SDS-PAGE
Samples were mixed with NuPAGE™ LDS sample buffer (4X) (Invitrogen, Waltham, MA, USA), loaded to a 4–20% pre-casted polyacrylamide gel (Biorad, USA), and analyzed under constant voltage of 200 V for 35 minutes. Proteins in the gel were stained by 1 hour incubation in InstantStain Coomassie Stain (INST-1L-181, Kem-En-Tec Nordic A/S, Denmark), followed by overnight destaining in dH2O. The concentration of αSyn was determined by densitometric analysis of the gels by ChemiDoc Go (BioRad, USA) using the Image Lab software (BioRad, USA) from the samples of known αSyn concentrations run in parallel as calibration.Intact mass spectroscopy of depolymerized fibrils
Fibrils were spun down as described earlier and the supernatant was removed. Solution of 8 M urea in 10 mM NaP buffer, pH 7.4 was added to depolymerize fibrils and samples were incubated for overnight (∼14 hours) at room temperature. Subsequently, the buffer was exchanged to 10 mM NaP buffer, pH 7.4 to remove urea from the sample. Samples were analyzed on an Ultraflex II MALDI-TOF/TOF Matrix-Assisted Laser Desorption Ionization (MALDI) mass spectrometer (Bruker). Samples for intact mass analysis were prepared for spotting by mixing 0.5 µL of sample with 0.5 µL of α-cyano-4-hydroxy-trans-cinnamic acid (saturated solution in 70% acetonitrile, 0.1% TFA). In addition to external calibration, α-Syn (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 Da) sample was used as an internal standard to improve accuracy.
460 Da) sample was used as an internal standard to improve accuracy.
Gel bands MS data acquisition and analysis
SDS-PAGE gels were washed in water and bands for analysis were excised and cut into pieces of 1 mm3 using a scalpel. Bands were destained for 30 min with constant mixing in 50% acetonitrile in 50 mM ammonium bicarbonate (AMBIC). After all color was removed, gel pieces were dehydrated by adding 500 µL 100% acetonitrile. Liquid was discarded and gel pieces were rehydrated in a solution of 10 mM TCEP, 40 mM CAA and 80 mM AMBIC. Gel pieces were incubated 30 min with constant shaking. 500 mM acetonitrile was added to dehydrate the gel pieces and excess liquid was removed. Leftover liquid was evaporated in a sterile bench. A solution of 25 ng µL−1 trypsin (MS grade, Sigma) in 10 mM AMBIC and 10% acetonitrile was added, and samples were incubated at 37 °C overnight with constant shaking. 50 µL of extraction buffer (5% formic acid in 50% acetonitrile) was added and samples incubated at 37 °C with constant shaking for 30 min. Supernatants containing the peptides were transferred to clean Lo-Bind Eppendorf tubes. Extraction was repeated and the pooled supernatants were concentrated in an Eppendorf Speedvac and re-constituted in 12 µL 2% acetonitrile, 1% TFA containing iRT peptides (Biognosys). Peptides were centrifuged at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10 min.
000×g for 10 min.
200 ng of the peptide mixture from each sample was analyzed by online nano-scale liquid chromatography tandem mass spectrometry (LC-MS/MS) in turn. Peptides were separated on a 15 cm C18-column (Thermo EasySpray ES804A) using an EASY-nLC 1200 system (Thermo Scientific). The column temperature was maintained at 30 °C. Buffer A consisted of 0.1% formic acid in water, and buffer B of 80% ACN, 0.1% formic acid. The flow rate of the gradient was kept at 250 nl min−1, and started at 6% buffer B, going to 23% buffer B in 20 min. This was followed by a 10 min step going to 38% buffer B, increasing to 60% buffer B in 3 minutes, and finally ramping up to 95% buffer B in 5 min, holding it for 7 min to wash the column. The Q Exactive Classic instrument (Thermo Scientific, Bremen, Germany) was run in data dependent acquisition mode using a top 10 Higher-energy Collisional Dissociation (HCD)-MS/MS method with the following settings. The scan range was limited to 350–1750 m/z. Full scan resolution was set to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m/z, with an AGC target of 3 × 106 and a maximum injection time (IT) value of 20 ms. Peptides were fragmented with a normalized collision energy of 25, having a dynamic exclusion of 30 s, excluding unassigned ions and those with a charge state of 1. MS/MS resolution was set at 17
000 m/z, with an AGC target of 3 × 106 and a maximum injection time (IT) value of 20 ms. Peptides were fragmented with a normalized collision energy of 25, having a dynamic exclusion of 30 s, excluding unassigned ions and those with a charge state of 1. MS/MS resolution was set at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 m/z, with an AGC target of 1 × 106 and a maximum IT of 60 ms.
500 m/z, with an AGC target of 1 × 106 and a maximum IT of 60 ms.
All raw LC-MS/MS data files were processed together using Proteome Discoverer version 2.4 (Thermo). In the processing step, oxidation, deamidation, protein N-termini acetylation and met-loss were set as dynamic modifications, with cysteine carbamidomethyl set as static modification. SequestHT was used as a database, matching spectra against the E. coli database and the P37840 entry from Uniprot.
Flow-induced dispersion analysis
The oligomeric state analysis of the supernatant and monomer quantification were further determined using the FIDA1 instrument (FidaBio, Denmark). Samples were analyzed using the following method:(1) Wash 1 (1 M NaOH): 45 s, 3500 mbar.
(2) Wash 2 (MQ water): 45 s, 3500 mbar.
(3) Equilibration (buffer): 30 s, 3500 mbar.
(4) Sample application (protein): 20 s, 75 mbar.
(5) Measurement and detection (buffer): 75 s, 1500 mbar.
All FIDA experiments were performed at 25 °C. The monomer concentration was quantified from the areas under the peak (obtained by fitting a Gaussian function to the Taylorgrams) using a calibration curve of known monomer concentrations.
Thioflavin T sensitivity assay
The sensitivity of the individual samples to Thioflavin T (ThT) was measured in 384-well low volume non-binding assay plates (Corning 3544) using a VANTAstar plate reader (BMG, Germany). A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution series of four samples was prepared for each aliquot using the respective buffer and supplemented with 50 µM ThT from a concentrated stock solution (5.5 mM). Each Fluorescence spectrum was measured in the 465–600 nm wavelength range upon excitation at 440 nm in 1 nm increments. The maximal intensity was plotted against fibril concentration calculated from the residual monomer analysis. Each sample was measured in duplicates (2 × 15 µL).
1 dilution series of four samples was prepared for each aliquot using the respective buffer and supplemented with 50 µM ThT from a concentrated stock solution (5.5 mM). Each Fluorescence spectrum was measured in the 465–600 nm wavelength range upon excitation at 440 nm in 1 nm increments. The maximal intensity was plotted against fibril concentration calculated from the residual monomer analysis. Each sample was measured in duplicates (2 × 15 µL).
Atomic force microscopy
All samples were diluted by the appropriate buffer to 5 µM monomer equivalent concentration (calculated by subtracting the residual monomer concentration from the total concentration) and 25 µL of the solution was deposited onto freshly cleaved mica substrates. After 2 min of incubation, the substrates were extensively cleaned by miliQ water and dried under nitrogen gas flow. All fibrils were imaged in tapping mode in air using a DriveAFM (Nanosurf, Liestal, Switzerland) using PPP-NCLAuD cantilevers (Nanosensors, Neuchatel, Switzerland). The AFM images were processed using the Gwyddion software. The apparent pitch length and height analysis of fibrils was carried out from the manually extracted fibril profiles using an automated python script.44,87 The script uses Fast Fourier Transform to detect frequencies of the z-axis data along the central ridge of the fibril which are then converted to the apparent pitch lengths (360° rotations) assuming a 21 screw axis symmetry.87 The assumption was made based on the structures of polymorhps 2a and 2b found to be formed in condition Fm.65Fibril sonication
A 200 µL aliquot was taken from each sample and centrifuged (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 90 min). Most (180 µL) of the supernatant was carefully removed and the pellets resuspended in their respective buffer to a monomer-equivalent concentration of 200 µM. Resuspended fibrils were sonicated using an ultrasonic probe (Hielscher UP200 St, Germany) in 3 s-pulses of 20% amplitude separated by 12 s pauses for 5 minutes (one minute total sonication time).
000×g, 90 min). Most (180 µL) of the supernatant was carefully removed and the pellets resuspended in their respective buffer to a monomer-equivalent concentration of 200 µM. Resuspended fibrils were sonicated using an ultrasonic probe (Hielscher UP200 St, Germany) in 3 s-pulses of 20% amplitude separated by 12 s pauses for 5 minutes (one minute total sonication time).
Circular dichroism
Sonicated fibrils were diluted to 15 µM and their circular dichroism (CD) spectra were recorded using a J-1500 CD spectrometer (Jasco, Japan). The spectra were collected in the 190–250 nm wavelength range at 50 nm min−1 scan rate and 0.5 nm steps at 25 °C. The final reported spectra are an average from triplicate measurements. The CD spectra of each buffer and αSyn monomer were collected as a reference. The CD spectrum of each fibril sample was deconvoluted using BeStSel webserver.88 The secondary structure content obtained from the analysis was averaged for each fibril type across all time points and repeats.The PCA (principal component analysis) of the CD spectra normalized by their respective areas (sum of absolute differences between spectrum and the zero baseline) was carried out using the PCA implementation from scikit-learn.89
Limited proteolysis
Fibrils were spun down as described earlier and the supernatant was removed. 20 mM Tris-HCl, pH 7.5, 1 mM EDTA was used to resuspend the pellet. Fibrils were sonicated (1 s on, 1 s off, 100% amplitude, 1 min total sonication time using an ultrasonic probe (Hielscher UP200 St, Germany)). 80 µM of fibrils were treated with 130 nM of proteinase K. At 0, 1, 5, 15 and 60 min, a sample was taken and mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with at 95 °C preheated NuPAGE™ LDS Sample Buffer (4X) (Thermo Fisher, USA). The samples were heated at 95 °C for 10 min to stop the reaction. The samples were analyzed by SDS-PAGE (NuPAGE™ bis–tris mini protein gels, 4–12%, Thermo Fisher, USA), run at a constant voltage of 150 V for 1 h. Color marker ultra-low range (#C6210, Merck KGaA, Darmstadt, Germany) was used as a protein standard. The gels were stained overnight in InstantStain Coomassie Stain (INST-1L-181, Kem-En-Tec Nordic A/S, Denmark) and destained with water. Lane profiles were extracted using ImageJ. The intensity at t = 0 min of the monomer band was set to 1 and all time points of the sample were normalized to this intensity.
1 with at 95 °C preheated NuPAGE™ LDS Sample Buffer (4X) (Thermo Fisher, USA). The samples were heated at 95 °C for 10 min to stop the reaction. The samples were analyzed by SDS-PAGE (NuPAGE™ bis–tris mini protein gels, 4–12%, Thermo Fisher, USA), run at a constant voltage of 150 V for 1 h. Color marker ultra-low range (#C6210, Merck KGaA, Darmstadt, Germany) was used as a protein standard. The gels were stained overnight in InstantStain Coomassie Stain (INST-1L-181, Kem-En-Tec Nordic A/S, Denmark) and destained with water. Lane profiles were extracted using ImageJ. The intensity at t = 0 min of the monomer band was set to 1 and all time points of the sample were normalized to this intensity.
Western blotting
SDS-PAGE gels were transferred onto 0.2 µm PVDF membranes (Trans-Blot Turbo Mini 0.2 µm PVDF Transfer Packs, Bio-Rad, Hercules, CA, USA) at 2.5 A for 7 min. Membranes were blocked in 5% milk in tris-buffered saline with Tween20 (TBST) at room temperature and subsequently washed three times in TBST for 5 min. The membranes were incubated with primary antibody solution (alpha Synuclein Antibody (3H2897): sc-69977 (Santa Cruz Biotechnology, Dallas, TX, USA), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 in 3% BSA in TBST, 0.02% sodium azide) over night at 4 °C. On the next day, the membranes were washed three times in TBST for 5 min. The secondary antibody solution (1
5000 in 3% BSA in TBST, 0.02% sodium azide) over night at 4 °C. On the next day, the membranes were washed three times in TBST for 5 min. The secondary antibody solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8000 Goat Anti-Mouse IgG, Human ads-HRP (#1030–05, Southern Biotech, Birmingham, AL, USA) in TBST) was incubated with the membranes for 1 h at room temperature. After washing the membranes three times in TBST for 5 min, the activity of the horse radish peroxidase was measured using Clarity™ Western ECL Substrate (#1705060, Bio-Rad, Hercules, CA, USA) and ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA).
8000 Goat Anti-Mouse IgG, Human ads-HRP (#1030–05, Southern Biotech, Birmingham, AL, USA) in TBST) was incubated with the membranes for 1 h at room temperature. After washing the membranes three times in TBST for 5 min, the activity of the horse radish peroxidase was measured using Clarity™ Western ECL Substrate (#1705060, Bio-Rad, Hercules, CA, USA) and ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA).
Thioflavin T seeding assay
The seeding potency of each fibril sample was measured in 384-well low volume non-binding assay plates (Corning 3544) using a FLUOstar plate reader (BMG, Germany). The monomer was diluted into disaggregation assay buffer (50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT) to final concentrations of 5, 10, 20, and 40 µM and supplemented with ThT (50 µM final concentration). Fixed volumes of sonicated fibrils (2.5 µM concentration, or the highest possible for the samples with low fibril concentration) were added to each sample and ThT fluorescence upon excitation at 440 nm was monitored at 480 nm under quiescent conditions in 5 min intervals at 37 °C. The initial parts of the curves (the first 2.5 hours) were fitted to a linear function, and the resulting slopes plotted against the initial monomer concentrations. The slope of the resulting curve corresponds to the apparent elongation rate (kapp) which is a product of the elongation rate constant (k+) and concentration of seeding-competent ends.The seeded assay with soluble fractions (supernatants after centrifugation) was carried out in the same way. A dilution series was made from each supernatant and supplemented with freshly made seeds (condition Fm, 2 weeks incubation at 37 °C, 600 rpm shaking, 2.5 µM final concentration) and ThT (50 µM final concentration). A dilution series of αSyn monomer buffer exchanged into each condition were prepared and used in the same way as a reference. The initial slope analysis was carried out as described above.
αSyn seed amplification assay (αSyn-SAA)
The αSyn-SAA was carried out as previously described.90,91 In brief, monomeric C-terminal His-tagged αSyn was mixed in 100 mM PIPES, pH 6.5, 500 mM NaCl, and 5 µM ThT at 1 mg mL−1 with brain tissue homogenates diluted to 10−4 in 1× PBS. The samples were subjected to cyclic agitation (1 min at 500 rpm followed by 29 min rest) at 37 °C. The ThT emission was monitored at 485 nm periodically using the Gemini-EM (Molecular Devices, CA, USA). The fibrils were harvested at the endpoint of the reaction, corresponding to 192 hours.Human samples
Human postmortem brain tissue from the pons regions from MSA and PD were kindly provided by the Bispebjerg Brain Bank, Centre for Neuroscience and Stereology, Department of Neurology, Copenhagen University Hospital, Bispebjerg and Frederiksberg Hospital, Denmark. The use of brain material from the Bispebjerg Brain Bank was approved under the regional ethical committee (j.no.: H-15016232) and Danish data protection agency (j.no.: P-2020-937).Urea depolymerization
Sonicated fibrils were diluted into 1× assay buffer (50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT) containing 0 to 4.8 M urea (40 µM final concentration in monomer-equivalents). Samples were incubated at room temperature for 3 days to allow for equilibration at the respective urea concentrations. After the incubation, the samples were analyzed by FIDA using the method described above. The monomer concentration was extracted from the elution profiles after correction for the viscosity at different urea concentrations as previously described.44 The chemical depolymerization curves were analyzed using NumPyro to sample posterior distributions of the isodesmic model (eqn (1))69 parameters using the No U-Turn Sampler (NUTS).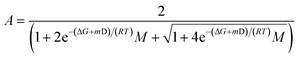 | (1) |
Data was analyzed using flat priors to visualize the maximum-likelihood estimation (MLE) or with a Gaussian prior on the m-values (denaturant dependence of thermodynamic stability) for a Bayesian treatment. The prior was based on its relationship to changes in solvent accessible area (ΔSASA) between structured (i.e., fibrillar) and disordered state (i.e., monomeric), derived from analysis of globular protein unfolding.70 The SASA values for the αSyn monomers in the fibrillar states were calculated from 13 representative structures of αSyn fibrils (pdb IDs 2n0a, 6a6b, 6cu7, 6cu8, 6osm, 6sst, 6ssx, 7nck, 7yk2, 7ynm, 8bqv, 8pk2, 9euu) using the FreeSASA python module.92 Only the SASA of a single monomer chain in the outermost layer of the fibrils was considered. The SASA of soluble, disordered state of αSyn monomers was calculated as a sum of per residue SASA (rSASA) contributions of residues buried in the corresponding fibril cores using average of different rSASA scales reviewed in.93,94
Amyloid fibrils amplified from brains of PD and MSA patients were analyzed by SDS-PAGE. Fibrils were sonicated and diluted into 1× assay buffer (50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT) containing 0 to 5.6 M urea (20 µM final concentration in monomer-equivalents). Samples were incubated at room temperature for 4 days to allow for equilibration at the respective urea concentrations. After the incubation, samples were centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900×g for 90 min at 25 °C. Supernatants were analyzed by SDS-PAGE as above. Band intensities were extracted using ImageLab. The analysis of the thermodynamic stability was performed as described above with a uniform prior for the m-value.
900×g for 90 min at 25 °C. Supernatants were analyzed by SDS-PAGE as above. Band intensities were extracted using ImageLab. The analysis of the thermodynamic stability was performed as described above with a uniform prior for the m-value.
Chaperone disaggregation
α-Synuclein fibrils were disaggregated by the human chaperone system Hsp70 + DNAJB1 + Apg2.52 The fibrils were centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g at 25 °C for 90 min in a tabletop centrifuge. The supernatant was discarded. The pellet was resuspended in the respective fibril buffer to a monomer-equivalent concentration of 200 µM. Fibrils were sonicated (1 min on, 1 min off, 1 min total sonication time, amplitude: 30%) using an ultrasonic probe (Hielscher UP200 St, Teltow, Germany). Fibrils were disaggregated at 2 µM by 4 µM Hsp70, 2 µM DNAJB1 and 0.2 µM Apg2 in 50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT and 2 mM ATP at 30 °C. The disaggregation was followed by 20 µM ThT with a VANTAstar plate reader (BMG LABTECH, Ortenberg, Germany) with a 440–10 nm excitation and 480–10 nm emission filter or a Fluostar plate reader (BMG LABTECH, Ortenberg, Germany). Samples were measured every 5 min with orbital shaking for 3 s before each cycle in a 384-well plate (Corning 3544).
000×g at 25 °C for 90 min in a tabletop centrifuge. The supernatant was discarded. The pellet was resuspended in the respective fibril buffer to a monomer-equivalent concentration of 200 µM. Fibrils were sonicated (1 min on, 1 min off, 1 min total sonication time, amplitude: 30%) using an ultrasonic probe (Hielscher UP200 St, Teltow, Germany). Fibrils were disaggregated at 2 µM by 4 µM Hsp70, 2 µM DNAJB1 and 0.2 µM Apg2 in 50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT and 2 mM ATP at 30 °C. The disaggregation was followed by 20 µM ThT with a VANTAstar plate reader (BMG LABTECH, Ortenberg, Germany) with a 440–10 nm excitation and 480–10 nm emission filter or a Fluostar plate reader (BMG LABTECH, Ortenberg, Germany). Samples were measured every 5 min with orbital shaking for 3 s before each cycle in a 384-well plate (Corning 3544).
Author contributions
A. K. B. supervised the work. C. F., A. K., A. S. W., B. B. and A. K. B. conceptualized the work. C. F. and A. K. designed and carried out the experiments, analyzed the data, prepared the graphics, and wrote the manuscript. S. W. performed the mass spectrometry experiments. R. K. N. and C. F. wrote the python code for the analysis of urea depolymerization experiments. M. S. and C. S participated in the seed amplification assay studies. T. L. D. guided chaperone mediated disaggregation experiments. All authors contributed to the preparation of the manuscript and agree with its content.Conflicts of interest
C. S. is a Founder, Chief Scientific Officer, consultant and shareholder of Amprion Inc., a biotechnology company that focuses on the commercial use of seed amplification assays for high-sensitivity detection of misfolded protein aggregates involved in various neurodegenerative diseases. The University of Texas Health Science Center at Houston has licensed patents and patent applications to Amprion.Data availability
Data for this article, including raw data of all experiments is available at Zenodo at https://doi.org/10.5281/zenodo.15212876.Supplementary information: detailed experimental characterization of fibril polymorphism, supplementary figures, and tables. See DOI: https://doi.org/10.1039/d5sc04927j.
Acknowledgements
A. K. B thanks the Novo Nordisk Foundation for funding (NNF17SA0028392 and NNF21OC0065495). This research was co-funded by the European Union (ERC CoG 101088163 EMMA to A.K.B.), Lundbeck foundation (grant number R366-2021-169 STADIC to A.K.B.), and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – project number 504257241 to BB. A. K. would like to acknowledge support through a Horizon MSCA individual postdoctoral fellowship (grant number 101106115) for funding. C. S. is supported by NIH grant U24AG079685 and M. S. by R01NS119689.References
- O. Sheppard and M. Coleman, Alzheimer's Disease: Etiology, Neuropathology and Pathogenesis, in Alzheimer's Disease: Drug Discovery, ed. Huang, X., Exon Publications, 2020, pp. 1–22 Search PubMed.
- A. Kouli, K. M. Torsney and W.-L. Kuan, Parkinson's Disease: Etiology, Neuropathology, and Pathogenesis. in Parkinson's Disease: Pathogenesis and Clinical Aspects, ed. Stoker, T. B. and Greenland, J. C., Codon Publications, 2018, pp. 3–26 Search PubMed.
- F. Chiti and C. M. Dobson, Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade, Annu. Rev. Biochem., 2017, 86, 27–68 CrossRef CAS PubMed.
- F. U. Hartl, Protein Misfolding Diseases, Annu. Rev. Biochem., 2017, 86, 21–26 CrossRef CAS.
- O. L. Lopez, et al., Association Between β-Amyloid Accumulation and Incident Dementia in Individuals 80 Years or Older Without Dementia, Neurology, 2024, 102, e207920 CrossRef CAS PubMed.
- A. Sturchio, et al., High cerebrospinal amyloid-β 42 is associated with normal cognition in individuals with brain amyloidosis, eClinicalMedicine, 2021, 38, 100988 CrossRef.
- G. P. Morris, I. A. Clark and B. Vissel, Inconsistencies and Controversies Surrounding the Amyloid Hypothesis of Alzheimer's Disease, Acta Neuropathol. Commun., 2014, 2, 135 Search PubMed.
- Y. C. Wong and D. Krainc, α-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies, Nat. Med., 2017, 23, 1–13 CrossRef CAS PubMed.
- H. A. Lashuel, C. R. Overk, A. Oueslati and E. Masliah, The many faces of α-synuclein: from structure and toxicity to therapeutic target, Nat. Rev. Neurosci., 2013, 14, 38–48 CrossRef CAS PubMed.
- M. K. Siddiqi, S. Malik, N. Majid, P. Alam and R. H. Khan, Cytotoxic species in amyloid-associated diseases: Oligomers or mature fibrils, in Advances in Protein Chemistry and Structural Biology, Elsevier, 2019, vol. 118, pp. 333–369 Search PubMed.
- Z.-L. Chen, P. K. Singh, M. Calvano, E. H. Norris and S. Strickland, A possible mechanism for the enhanced toxicity of beta-amyloid protofibrils in Alzheimer's disease, Proc. Natl. Acad. Sci. U. S. A, 2023, 120, e2309389120 CrossRef CAS.
- Y. Shi, et al., Structure-based classification of tauopathies, Nature, 2021, 598, 359–363 CrossRef CAS.
- R. W. L. So and J. C. Watts, α-Synuclein Conformational Strains as Drivers of Phenotypic Heterogeneity in Neurodegenerative Diseases, J. Mol. Biol., 2023, 435, 168011 CrossRef CAS PubMed.
- Y. Yang, et al., Structures of α-synuclein filaments from human brains with Lewy pathology, Nature, 2022, 610, 791–795 CrossRef CAS.
- R. Tycko, Amyloid Polymorphism: Structural Basis and Neurobiological Relevance, Neuron, 2015, 86, 632–645 CrossRef CAS.
- M. G. Iadanza, M. P. Jackson, E. W. Hewitt, N. A. Ranson and S. E. Radford, A new era for understanding amyloid structures and disease, Nat. Rev. Mol. Cell Biol., 2018, 19, 755–773 CrossRef CAS.
- E. Chatani, K. Yuzu, Y. Ohhashi and Y. Goto, Current Understanding of the Structure, Stability and Dynamic Properties of Amyloid Fibrils, Int. J. Mol. Sci., 2021, 22, 4349 CrossRef CAS PubMed.
- M. R. Sawaya, M. P. Hughes, J. A. Rodriguez, R. Riek and D. S. Eisenberg, The expanding amyloid family: Structure, stability, function, and pathogenesis, Cell, 2021, 184, 4857–4873 CrossRef CAS PubMed.
- M. D. Gelenter, et al., The peptide hormone glucagon forms amyloid fibrils with two coexisting β-strand conformations, Nat. Struct. Mol. Biol., 2019, 26, 592–598 CrossRef CAS PubMed.
- C. B. Andersen, et al., Glucagon Fibril Polymorphism Reflects Differences in Protofilament Backbone Structure, J. Mol. Biol., 2010, 397, 932–946 CrossRef CAS.
- J. S. Pedersen, et al., The Changing Face of Glucagon Fibrillation: Structural Polymorphism and Conformational Imprinting, J. Mol. Biol., 2006, 355, 501–523 CrossRef CAS.
- A. Sakalauskas, M. Ziaunys and V. Smirnovas, Concentration-dependent polymorphism of insulin amyloid fibrils, PeerJ, 2019, 7, e8208 CrossRef.
- S. Suladze, et al., Atomic resolution structure of full-length human insulin fibrils, Proc. Natl. Acad. Sci. U. S. A, 2024, 121, e2401458121 CrossRef CAS.
- M. Ziaunys, T. Sneideris and V. Smirnovas, Formation of distinct prion protein amyloid fibrils under identical experimental conditions, Sci. Rep., 2020, 10, 4572 CrossRef CAS.
- M. Ziaunys, A. Sakalauskas, K. Mikalauskaite and V. Smirnovas, Polymorphism of Alpha-Synuclein Amyloid Fibrils Depends on Ionic Strength and Protein Concentration, Int. J. Mol. Sci., 2021, 22, 12382 CrossRef CAS PubMed.
- W. Hoyer, et al., Dependence of α-Synuclein Aggregate Morphology on Solution Conditions, J. Mol. Biol., 2002, 322, 383–393 CrossRef CAS.
- M. Ziaunys, A. Sakalauskas and V. Smirnovas, Identifying Insulin Fibril Conformational Differences by Thioflavin-T Binding Characteristics, Biomacromolecules, 2020, 21, 4989–4997 Search PubMed.
- D. R. Boyer, et al., Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs, Nat. Struct. Mol. Biol., 2019, 26, 1044–1052 CrossRef CAS PubMed.
- C. Sun, et al., Cryo-EM structure of amyloid fibril formed by α-synuclein hereditary A53E mutation reveals a distinct protofilament interface, J. Biol. Chem., 2023, 299, 104566 CrossRef CAS PubMed.
- X. Ni, R. P. McGlinchey, J. Jiang and J. C. Lee, Structural Insights into α-Synuclein Fibril Polymorphism: Effects of Parkinson's Disease-Related C-Terminal Truncations, J. Mol. Biol., 2019, 431, 3913–3919 Search PubMed.
- K. Zhao, et al., Parkinson's disease-related phosphorylation at Tyr39 rearranges α-synuclein amyloid fibril structure revealed by cryo-EM, Proc. Natl. Acad. Sci. U. S. A, 2020, 117, 20305–20315 CrossRef CAS PubMed.
- A. T. Balana, et al., O-GlcNAc forces an α-synuclein amyloid strain with notably diminished seeding and pathology, Nat. Chem. Biol., 2024, 20, 646–655 CrossRef CAS PubMed.
- D. Liu, et al., Differential seeding and propagating efficiency of α-synuclein strains generated in different conditions, Transl. Neurodegener., 2021, 10, 20 CrossRef CAS.
- L. Bousset, et al., Structural and functional characterization of two alpha-synuclein strains, Nat. Commun., 2013, 4, 2575 CrossRef PubMed.
- A. Sidhu, I. Segers-Nolten, V. Raussens, M. M. A. E. Claessens and V. Subramaniam, Distinct Mechanisms Determine α-Synuclein Fibril Morphology during Growth and Maturation, ACS Chem. Neurosci., 2017, 8, 538–547 CrossRef CAS.
- Y. Hou, et al., Ageing as a risk factor for neurodegenerative disease, Nat. Rev. Neurol., 2019, 15, 565–581 Search PubMed.
- A. Miller, et al., Maturation-dependent changes in the size, structure and seeding capacity of Aβ42 amyloid fibrils, Commun. Biol., 2024, 7, 153 CrossRef CAS PubMed.
- S. Becker, K. Giller, D. Sieme and N. Rezaei-Ghaleh, Maturation of amyloid β fibrils alters their molecular stability, Phys. Chem. Chem. Phys., 2023, 25, 15099–15103 RSC.
- S. Mehra, et al., α-Synuclein Aggregation Intermediates form Fibril Polymorphs with Distinct Prion-like Properties, J. Mol. Biol., 2022, 434, 167761 CrossRef CAS PubMed.
- T. Pálmadóttir, et al., Morphology-Dependent Interactions between α-Synuclein Monomers and Fibrils, Int. J. Mol. Sci., 2023, 24, 5191 CrossRef.
- S. Lövestam, et al., Disease-specific tau filaments assemble via polymorphic intermediates, Nature, 2024, 625, 119–125 CrossRef PubMed.
- M. Wilkinson, et al., Structural evolution of fibril polymorphs during amyloid assembly, Cell, 2023, 186, 5798–5811 CrossRef CAS PubMed.
- A. J. Baldwin, et al., Metastability of Native Proteins and the Phenomenon of Amyloid Formation, J. Am. Chem. Soc., 2011, 133, 14160–14163 CrossRef CAS.
- A. Farzadfard and A. Kunka, et al., Thermodynamic characterization of amyloid polymorphism by microfluidic transient incomplete separation, Chem. Sci., 2024, 15, 2528–2544 RSC.
- H. H. Kampinga and S. Bergink, Heat shock proteins as potential targets for protective strategies in neurodegeneration, Lancet Neurol., 2016, 15, 748–759 CrossRef CAS.
- J. Labbadia and R. I. Morimoto, The Biology of Proteostasis in Aging and Disease, Annu. Rev. Biochem., 2015, 84, 435–464 CrossRef CAS.
- A. Wentink, C. Nussbaum-Krammer and B. Bukau, Modulation of Amyloid States by Molecular Chaperones, Cold Spring Harbor Perspect. Biol., 2019, 11, a033969 CrossRef CAS.
- C. Huang, P. Rossi, T. Saio and C. G. Kalodimos, Structural basis for the antifolding activity of a molecular chaperone, Nature, 2016, 537, 202–206 CrossRef CAS PubMed.
- A. Ciechanover and Y. T. Kwon, Protein Quality Control by Molecular Chaperones in Neurodegeneration, Front. Neurosci., 2017, 11, 1–18 Search PubMed.
- Z. L. Almeida and R. M. M. Brito, Amyloid Disassembly: What Can We Learn from Chaperones?, Biomedicines, 2022, 10, 3276 CrossRef CAS PubMed.
- H. Saibil, Chaperone machines for protein folding, unfolding and disaggregation, Nat. Rev. Mol. Cell Biol., 2013, 14, 630–642 CrossRef CAS PubMed.
- A. S. Wentink, et al., Molecular dissection of amyloid disaggregation by human HSP70, Nature, 2020, 587, 483–488 CrossRef CAS PubMed.
- M. L. Duennwald, A. Echeverria and J. Shorter, Small Heat Shock Proteins Potentiate Amyloid Dissolution by Protein Disaggregases from Yeast and Humans, PLoS Biol., 2012, 10, e1001346 CrossRef CAS PubMed.
- X. Gao, et al., Human Hsp70 Disaggregase Reverses Parkinson’s-Linked α-Synuclein Amyloid Fibrils, Mol. Cell, 2015, 59, 781–793 CrossRef CAS.
- M. M. Schneider, et al., The Hsc70 disaggregation machinery removes monomer units directly from α-synuclein fibril ends, Nat. Commun., 2021, 12, 5999 CrossRef CAS.
- E. Nachman, et al., Disassembly of Tau fibrils by the human Hsp70 disaggregation machinery generates small seeding-competent species, J. Biol. Chem., 2020, 295, 9676–9690 CrossRef CAS.
- A. Scior, et al., Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex, EMBO J., 2018, 37, 282–299 CrossRef CAS PubMed.
- P. Goloubinoff and P. D. L. Rios, The mechanism of Hsp70 chaperones: (entropic) pulling the models together, Trends Biochem. Sci., 2007, 32, 372–380 CrossRef CAS PubMed.
- R. Sousa and E. M. Lafer, The Physics of Entropic Pulling: A Novel Model for the Hsp70 Motor Mechanism, Int. J. Mol. Sci., 2019, 20, 2334 CrossRef CAS.
- P. De Los Rios, A. Ben-Zvi, O. Slutsky, A. Azem and P. Goloubinoff, Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling, Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 6166–6171 CrossRef CAS PubMed.
- S. Jäger, et al., Structural polymorphism of α-synuclein fibrils alters pathway of Hsc70 mediated disaggregation, EMBO J., 2025, 1–28 Search PubMed.
- A. Makky, L. Bousset, J. Polesel-Maris and R. Melki, Nanomechanical properties of distinct fibrillar polymorphs of the protein α-synuclein, Sci. Rep., 2016, 6, 37970 CrossRef CAS PubMed.
- L. Bousset, et al., α-Synuclein Fibril, Ribbon and Fibril-91 Amyloid Polymorphs Generation for Structural Studies, in Protein Aggregation: Methods and Protocols, ed. Cieplak, A. S., Humana Press, New York, NY, 2023, vol. 2551 Search PubMed.
- J. Gath, et al., Solid-state NMR sequential assignments of α-synuclein, Biomol. NMR Assignments, 2012, 6, 51–55 CrossRef CAS.
- R. Guerrero-Ferreira, et al., Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy, eLife, 2019, 8, e48907 CrossRef CAS PubMed.
- J. Verasdonck, et al., Further exploration of the conformational space of α-synuclein fibrils: solid-state NMR assignment of a high-pH polymorph, Biomol. NMR Assignments, 2016, 10, 5–12 CrossRef CAS.
- N. Vettore and A. K. Buell, Thermodynamics of amyloid fibril formation from chemical depolymerization, Phys. Chem. Chem. Phys., 2019, 21, 26184–26194 RSC.
- T. Narimoto, et al., Conformational stability of amyloid fibrils of β2 -microglobulin probed by guanidine-hydrochloride-induced unfolding, FEBS Lett., 2004, 576, 313–319 CrossRef CAS PubMed.
- K. L. Callaghan, Thermodynamic Characterisation of Amyloid Fibrils, Apollo - University of Cambridge Repository, 2021, DOI:10.17863/CAM.82240.
- J. K. Myers, C. Nick Pace and J. Martin Scholtz, Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding, Protein Sci., 1995, 4, 2138–2148 CrossRef CAS.
- J. M. Scholtz, G. R. Grimsley and C. N. Pace, Chapter 23 Solvent Denaturation of Proteins and Interpretations of the m Value, Methods Enzymol., 2009, 466, 549–565 CAS.
- J. H. M. van Gils, et al., The hydrophobic effect characterises the thermodynamic signature of amyloid fibril growth, PLoS Comput. Biol., 2020, 16, e1007767 CrossRef CAS PubMed.
- L. Frey, et al., On the pH-dependence of α-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation, eLife, 2024, 12, RP93562 CrossRef.
- S. Lövestam, M. Schweighauser, T. Matsubara, S. Murayama, T. Tomita, T. Ando, K. Hasegawa, M. Yoshida, A. Tarutani, M. Hasegawa, M. Goedert and S. H. W. Scheres, Seeded assembly in vitro does not replicate the structures of α-synuclein filaments from multiple system atrophy, FEBS Open Bio, 2021, 11, 999–1013 CrossRef PubMed.
- M. Shahnawaz, A. Mukherjee and S. Pritzkow, et al., Discriminating α-synuclein strains in Parkinson's disease and multiple system atrophy, Nature, 2020, 578, 273–277 CrossRef CAS PubMed.
- A.-L. Mahul-Mellier, et al., The process of Lewy body formation, rather than simply α-synuclein fibrillization, is one of the major drivers of neurodegeneration, Proc. Natl. Acad. Sci. U. S. A, 2020, 117, 4971–4982 CrossRef CAS.
- V. Redeker, S. Pemberton, W. Bienvenut, L. Bousset and R. Melki, Identification of Protein Interfaces between α-Synuclein, the Principal Component of Lewy Bodies in Parkinson Disease, and the Molecular Chaperones Human Hsc70 and the Yeast Ssa1p, J. Biol. Chem., 2012, 287, 32630–32639 CrossRef CAS PubMed.
- B. M. Burmann, et al., Regulation of α-synuclein by chaperones in mammalian cells, Nature, 2020, 577, 127–132 CrossRef CAS PubMed.
- M. Landureau, V. Redeker, T. Bellande, S. Eyquem and R. Melki, The differential solvent exposure of N-terminal residues provides “fingerprints” of alpha-synuclein fibrillar polymorphs, J. Biol. Chem., 2021, 296, 100737 CrossRef CAS PubMed.
- A.-L. Mahul-Mellier, et al., Dissecting the differential role of C-terminal truncations in the regulation of aSyn pathology formation and the biogenesis of Lewy bodies, npj Parkinson's Dis., 2024 DOI:10.1101/2024.11.29.625993.
- C. Zhang, et al., C-terminal truncation modulates α-Synuclein’s cytotoxicity and aggregation by promoting the interactions with membrane and chaperone, Commun. Biol., 2022, 5, 798 CrossRef CAS PubMed.
- L. Ma, et al., C-terminal truncation exacerbates the aggregation and cytotoxicity of α-Synuclein: A vicious cycle in Parkinson's disease, Biochim. Biophys. Acta Mol. Basis Dis., 2018, 1864, 3714–3725 CrossRef CAS PubMed.
- G. Muntané, I. Ferrer and M. Martinez-Vicente, α-synuclein phosphorylation and truncation are normal events in the adult human brain, Neuroscience, 2012, 200, 106–119 CrossRef.
- R. Porcari, et al., The H50Q Mutation Induces a 10-fold Decrease in the Solubility of α-Synuclein, J. Biol. Chem., 2015, 290, 2395–2404 CrossRef CAS PubMed.
- J. Gath, et al., Unlike Twins: An NMR Comparison of Two α-Synuclein Polymorphs Featuring Different Toxicity, PLoS ONE, 2014, 9, e90659 CrossRef PubMed.
- R. C. Phillips, P. George and R. J. Rutman, Thermodynamic Data for the Hydrolysis of Adenosine Triphosphate as a Function of pH, Mg2+ Ion Concentration, and Ionic Strength, J. Biol. Chem., 1969, 244, 3330–3342 CrossRef CAS PubMed.
- J. A. Larsen, Mechanistic Studies of Amyloid Propagation, DTU, 2023 Search PubMed.
- A. Misconai, E. Moussong, F. Wien, E. Boros, H. Vadászi, N. Murvai, Y. Lee, T. Molnar, M. Réfrégiers, Y. Goto, A. Tantos and J. Kardos, BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy, Nucleic Acids Res., 2022, 50 W1, W90–W98 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, Scikit-learn: Machine Learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- M. Shahnawaz, T. Tokuda, M. Waragai, N. Mendez, R. Ishii, C. Trenkwalder, B. Mollenhauer and C. Soto, Development of a Biochemical Diagnosis of Parkinson Disease by Detection of α-Synuclein Misfolded Aggregates in Cerebrospinal Fluid, JAMA Neurol., 2017, 74(2), 163–172 CrossRef PubMed.
- M. Shahnawaz, A. Mukherjee and S. Pritzkow, et al., Discriminating α-synuclein strains in Parkinson's disease and multiple system atrophy, Nature, 2020, 578, 273–277 CrossRef CAS PubMed.
- S. Mitternacht, FreeSASA: An open source C library for solvent accessible surface area calculations, F1000Research, 2016, 5, 189 Search PubMed.
- S. Lu and A. S. Wagaman, On methods for determining solvent accessible surface area for proteins in their unfolded state, BMC Res. Notes, 2014, 7, 602 CrossRef PubMed.
- S. Ali, Md. Hassan, A. Islam and F. Ahmad, A Review of Methods Available to Estimate Solvent-Accessible Surface Areas of Soluble Proteins in the Folded and Unfolded States, Curr. Protein Pept. Sci., 2014, 15, 456–476 CrossRef CAS.
Footnote |
| † These authors contributed equally: Fricke C., Kunka A. |
| This journal is © The Royal Society of Chemistry 2026 |



