Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Continuous and sensitive monitoring of LPMO reactions using an optical H2O2 sensor
Leonor Vieira Carneiro ab,
Anders Ørts Tjellc,
María Dolores Fernández-Ramos
ab,
Anders Ørts Tjellc,
María Dolores Fernández-Ramos d,
Torsten Mayr
d,
Torsten Mayr *c and
Daniel Kracher
*c and
Daniel Kracher *ab
*ab
aInstitute of Molecular Biotechnology, Graz University of Technology, Petersgasse 14, 8010 Graz, Austria. E-mail: daniel.kracher@tugraz.at; Tel: +43 316 873 4084
bBioTechMed-Graz, Graz, Austria
cInstitute of Analytical Chemistry and Food Chemistry, Graz University of Technology, Stremayrgasse 9/II, 8010 Graz, Austria. E-mail: torsten.mayr@tugraz.at; Tel: +43 316 873 32504
dDepartment of Analytical Chemistry, University of Granada, 18071 Granada, Spain
First published on 4th December 2025
Abstract
We demonstrate the use of an optical sensor to follow the activity of H2O2-dependent enzymes in real-time, using quasi-continuous detection of H2O2. The sensor enables kinetic measurements of the H2O2-consuming enzyme lytic polysaccharide monooxygenase (LPMO) under homogeneous conditions at 1 mL scale conversion assays. By tracking H2O2 concentration changes over time during 2-minute reaction intervals, we established quantitative assays and determined Michaelis–Menten kinetics for the LPMO from Lentinus similis to cellotetraose, with a KM value of 0.22 ± 0.08 mM and a maximum reaction rate (Vmax) of 0.97 ± 0.64 μM s−1. This method allows for continuous monitoring without reliance on time-consuming, discontinuous assays or post-reaction sample processing. We evaluate the capabilities of the sensor to monitor enzyme activity, benchmark it against established spectrophotometric methods, and discuss its limitations and advantages. This work provides a method for the real-time assessment of LPMO kinetics but also enables broader application to other redox enzymes that consume or release H2O2 during the reaction.
1 Introduction
Redox enzymes such as peroxidases, peroxygenases and oxidases catalyse a wide range of oxidation and oxyfunctionalisation reactions using hydrogen peroxide (H2O2) or molecular oxygen (O2) as co-substrates.1–5 Among those, lytic polysaccharide monooxygenases are ubiquitous copper-dependent enzymes6 that employ a H2O2-dependent redox mechanism to cleave recalcitrant polysaccharides such as cellulose and chitin.7 LPMOs break β-1,4-glycosidic linkages in the substrate through oxidation at the C1 and/or C4 carbons, thereby enhancing the overall efficiency of cellulolytic enzymes.8–10 As a result, these enzymes can boost commercial cellulase mixtures in biorefinery settings.11LPMOs have been identified in a wide range of host organisms, including bacteria, fungi, plants and insects.12–14 Based on their substrate scope and sequence, they are classified into multiple auxiliary activity (AA) families in the CAZy database (AA9-11, AA13-15, and AA16-17; http://www.cazy.org). Originally thought to use O2 as a co-substrate,6 recent research has demonstrated that LPMOs possess a dominant peroxygenase reactivity that is several orders of magnitude higher than the reaction with molecular oxygen.7,15–18 While this reaction enables enhanced catalytic efficiency, it also increases the risk of oxidative self-inactivation under substrate-depleted conditions.7,19–21 The LPMO reaction requires not only externally supplied H2O2 but also reduction equivalents, which can be derived from redox partner enzymes or small molecule reductants, such as ascorbic acid.22
Activity determination of LPMOs typically relies on chromatographic methods that separate and quantify the diverse soluble oxidised oligosaccharides released from the polysaccharide substrate by LPMOs. This necessitates the availability of oxidised oligosaccharide standards and requires derivatisation or specialised detection techniques such as HPLC-MS/UHPLC-MS, HPAEC-PAD or MALDI-TOF. A detailed overview of chromatographic methods for the qualitative and quantitative analysis of LPMOs is provided elsewhere.23,24 While indispensable to determine the product profile and gain insight into the mechanistics of the reaction, chromatographic methods are generally not suitable for real-time monitoring of LPMO activity.
Several rapid assays have been developed to monitor LPMO activity using fluorogenic or chromogenic substrates in time-resolved formats.25 Among these, the Amplex™ Red assay detects the H2O2 generated by reduced LPMO through a slow oxygen-dependent uncoupling reaction, but is limited by sensitivity to light and air, and interference from reducing agents.26 Alternatively, LPMOs exhibit peroxidase-like activity, enabling spectrophotometric detection through H2O2-dependent oxidation of 2,6-dimethoxyphenol or hydrocoerulignone, forming coerulignone detectable at 469 nm.27,28 A highly sensitive fluorometric assay has also been developed based on LPMO-catalysed oxidation of reduced fluorescein to fluorescein.29 However, it is important to note that these assays, even though useful to study LPMO stability and functionality, do not provide direct information on the polysaccharide-dependent peroxygenase activity of LPMOs.
Discontinuous assays have also been established, including turbidity-based measurements of substrate depletion30 and quantification of oxidised products by Ni2+ adsorption.31 The Ni2+ adsorption assay measures C1-oxidation by detecting aldonic acid products through nickel ion binding.31 Also, gluconic acid produced by C1-oxidising LPMOs can be quantified through the concomitant formation of NADPH, which is then measured spectrophotometrically.32 However, these assays are limited to specific reaction conditions (turbidity assay), or require C1-oxidising LPMOs (Ni2+ assay, D-gluconic acid/D-glucono-δ-lactone assay kit). Additionally, a colourimetric assay using dyed azoxyloglucan detects activity through release of coloured oligosaccharides, though its general applicability is restricted to LPMOs active on xylan or its derived oligosaccharides.24
Alternatively, LPMO activity has also been measured via the determination of the depletion of the co-substrate H2O2. Accurate monitoring of enzyme activity via quantification of H2O2 is often complicated by its chemical instability and rapid reaction with reductants.5,33 Recently, electrochemical methods have been applied to detect H2O2-depletion in LPMO reactions, thus providing real-time kinetic information.17,34 A microsensor was used to detect the H2O2 consumption by fungal LPMOs acting on a native cellulose substrate (plant cell walls). The method provides high spatial resolution of H2O2 consumption but also requires a complex scanning electrochemical microscopy (SECM) platform.17 Alternatively, consumption of H2O2 by LPMO on soluble and insoluble substrates was monitored in real-time using a gold rotating disc electrode modified with a thin layer of Prussian blue to achieve selective detection of H2O2.34 This setup reduced diffusion limitations and allowed for a detailed insight into enzyme kinetics, stability, and substrate specificity of various tested LPMOs. While such electrochemical systems offer high selectivity due to the low applied potential (100 mV vs. SHE), they require expertise in electroanalytical techniques, specialised instrumentation and are challenging to implement for at-line monitoring of H2O2.
In this research, we employ an optical H2O2 sensor, first reported by Tjell et al.,35,36 to monitor the activity of H2O2-generating and consuming enzymes. The sensor was previously applied to follow H2O2 formation by nitrogen-doped carbon nanodots36 and to follow the enzymatically produced H2O2 by immobilised glucose oxidase in a model reactor.35 We show that this sensor concept can be extended to the measurement of freely diffusing enzymes. Importantly, its low response time in the seconds range, its straightforward setup and the broad dynamic range from a few micromolar to a few hundred micromolar, make it a useful tool in biocatalysis for mechanistic studies as well as for process monitoring. Furthermore, the sensor exhibits minimal cross-sensitivities to interfering substances, with responses relative to H2O2 measured as 2.7% for sodium peroxynitrite, 0.2% for sodium hypochlorite, 0.1% for tert-butyl hydroperoxide, and 2.2% for ascorbic acid (each species tested at 100 μM). Salinity (10 mM phosphate buffer vs. filtered seawater) did not affect the sensor calibration.35 We demonstrate its utility by following the activity of a H2O2-producing and a H2O2-consuming enzyme, glucose oxidase (GOD) and LPMO from Lentinus similis (LsAA9A), respectively. LsAA9A exhibits activity on both soluble oligosaccharides, as short as cellotriose, and insoluble polysaccharide substrates, including cellulose and xylan,37–41 making it an ideal candidate for evaluating H2O2-driven oxidative activity. By directly tracking the consumption of H2O2 by the LPMO, which is utilised stoichiometrically with the formation of oxidised products (e.g., one mole of H2O2 consumed per mole of oxidised sugar),7 the sensor allows for a rapid, substrate-independent monitoring of the activity of this enzyme.
2 Results and discussion
2.1 Set-up of H2O2 optical sensor
The H2O2 sensor used was first reported by Tjell et al.35 The setup used in these experiments (Fig. 1A and C) consists of luminescence-based sensing elements integrated into a flow cell. The sensing elements consist of a platinum-based catalyst and luminescent oxygen-sensitive particles embedded in a host polymer. The catalyst converts H2O2 in the sample solution into molecular oxygen (O2), which is detected and quantified by the oxygen-sensitive particles. To eliminate the background from dissolved O2 in the sample, the sample solution is pumped through sodium sulphite solution (Na2SO3, 2%) through a gas-permeable silicon tube to scavenge oxygen before entering the flow cell. Importantly, this avoids direct contact of the reaction mixture with Na2SO3. | ||
| Fig. 1 Optical H2O2 sensor setup35 and H2O2-dependent enzymatic reactions tested. (A) Schematic representation of the sensor setup. The reaction mixture passes through an oxygen scavenging solution of 2% Na2SO3 inside a gas-permeable but Na2SO3 impermeable silicone tube (l = 29 cm; OD = 2.5 mm) to scavenge atmospheric O2 in the sample mixture. The flow cell (l = 25 mm) contains two sensor spots; one functions as an O2 sensor, while the other includes the catalyst converting the H2O2 to O2. The O2 concentration is quantified through oxygen-sensitive particles. The H2O2 concentrations are calculated from the difference between both O2 signals recorded on two separate channels. (Created with http://BioRender.com) (B) glucose oxidase (GOD) and LsAA9A were used as H2O2-forming and H2O2-consuming enzymes, respectively, to test the optical sensor for monitoring of enzyme activity. GOD is an oxidase that oxidises glucose into D-glucono-1,5-lactone in the presence of O2. The LsAA9A cleaves both soluble and insoluble cello-oligosaccharides. Here we test the LsAA9A with soluble cellotetraose in the presence of H2O2 and ascorbic acid as reducing agent for the reduction of the copper active centre of the enzyme (AscA = ascorbic acid; dhAscA = dehydroascorbic acid). http://BioRender.com (C) setup of the prototype optical H2O2 sensor in the laboratory. A syringe pump is used to pump the reaction mixture through the sensor system. | ||
The optical H2O2 sensor was evaluated using the H2O2-producing enzyme glucose oxidase in a homogeneous reaction system.
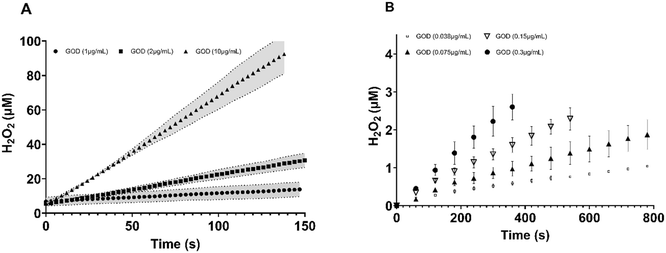
The optical H2O2 sensor enabled continuous, real-time measurement of H2O2 production in a concentration range of 5 μM to 200 μM.35 To evaluate the suitability of the herein used setup for enzyme assays, glucose oxidase (GOD) was employed as a model H2O2-producing enzyme in a homogeneous aqueous environment. GOD catalyses the oxidation of glucose to gluconic acid, with the concurrent formation of H2O2. In measurements with the optical H2O2 sensor (Fig. 2A), we used a temporal resolution of 3 seconds, allowing for a quasi-continuous monitoring of the enzymatic reaction. In these experiments, we obtained a specific GOD activity of 5.1 U genzyme−1, at pH 6 and 37 °C. To validate this result, an orthogonal spectrophotometric method using 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) as chromogenic substrate was employed using the same conditions. In this indirect method, ABTS is oxidised by horseradish peroxidase in the presence of H2O2 produced by GOD, producing a measurable colour change (Fig. 2B). Through this method, we obtained a specific GOD activity of 7.9 U genzyme−1. The discrepancy between the activity data, showing an approximately 1.5-times lower H2O2 formation rate of the optical sensor, can be explained by several factors. Notably, the photometric ABTS-coupled assay is limited to H2O2 concentrations below ∼5 μM and requires sub-μM concentrations of GOD due to the strong absorbance of the formed ABTS cation radical. In contrast, the optical sensor can reliably detect H2O2 concentrations up to 80 μM (Fig. S2), allowing for a broader measurement range. The lower response of the H2O2 sensor may also result from the inherent delay associated with the design of the flow system. Before entering the optical flow cell, around 100 μL of reaction mixture passes through a deoxygenation module containing sodium sulphite, which is essential for minimising background interference caused by dissolved oxygen. This delay may result in partial degradation or disproportionation of the enzymatically produced H2O2, thus reducing the concentration of O2 detected by the luminescence probe. Additionally, the internal catalytic activity of the flow cell and diffusion limitations might attenuate the sensor response further, especially during rapid enzymatic turnover. Despite this, these data demonstrate the general suitability of the sensor to measure enzyme reactions in which enzymes are freely diffusing.
2.2 Evaluation of optical H2O2 sensor using H2O2-degrading LPMO in homogeneous reaction systems
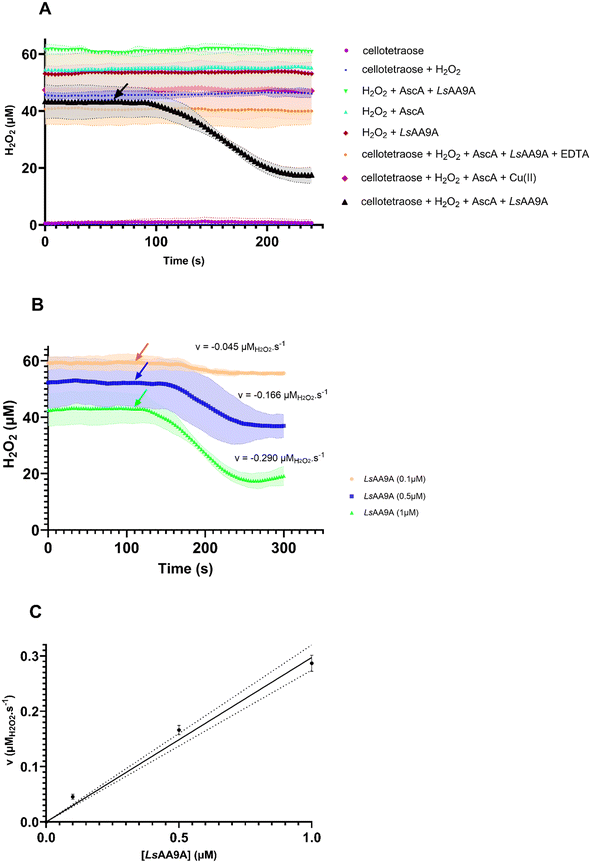
To assess H2O2 consumption, we selected an LPMO from Lentinus similis (LsAA9A), a well-studied LPMO38 exhibiting activity on both soluble oligosaccharides, as short as cellotriose, and insoluble polysaccharide substrates, including cellulose.37–41 In this study, LPMO reactions were performed using 50 μM ascorbic acid (AscA) as the reducing agent together with cellotetraose, a soluble oligosaccharide substrate (Fig. 3A). Initial H2O2 concentrations were set at 50 μM, which falls within the range of commonly reported concentrations for LPMO activity determinations (50–100 μM). This is notable because many LPMOs are known to tolerate H2O2 concentrations only up to ∼100 μM before the risk of autoxidative degradation becomes significant.42 With the H2O2 sensor, the initially measured H2O2 concentrations showed deviations of 10–20% (Fig. 3A).A series of control reactions were performed to distinguish background signals from enzymatic H2O2 consumption (Fig. 3A). Cellotetraose alone at a concentration of 0.1 mM did not interfere with the output signal, confirming the absence of the endogenous H2O2 and absence of interference caused by other contaminants present within the substrate solution. Reactions containing only ascorbic acid and H2O2, as well as those combining cellotetraose with H2O2 in the absence of enzyme, showed that none of the components individually or in combination interfered with the H2O2 detection. Additional controls included reactions containing only Cu(II), as well as enzyme preparations in which the copper cofactor had been chelated by EDTA (5000-fold molar excess over LsAA9A) to assess the role of the active-site metal centre. These controls demonstrated that H2O2 consumption only occurs in the copper-bound state of the enzyme and in the presence of all reaction components (Fig. 3A).
Additional reactions were conducted with or without ascorbic acid and cellotetraose. Control reactions containing LsAA9A and H2O2 in the absence of cellotetraose showed a slow decrease in the H2O2 concentration, possibly due to the release of copper upon enzyme deactivation.42 However, these reactions were at least an order of magnitude slower than the substrate-dependent turnover and not significant within the observed timeframe (data not shown).
Assays containing all reaction components were initiated by the addition of LsAA9A at varying concentrations (Fig. 3A and B). The enzyme was added after the stabilisation of the H2O2 signal. After a short lag phase, a decrease in H2O2 was observed, indicative of LPMO catalysis (Fig. 3A and B). Reaction rates were calculated from the initial slopes of the reactions shown in Fig. 3B. Notably, the rates of H2O2-depletion were proportional to the employed LsAA9A concentration (Fig. 3B and C). Additionally, samples were periodically withdrawn during the assay and oxidised products were quantified.32 The time-dependent formation of oxidised products corroborated the activity of the LsAA9A on the tetraose substrate (Fig. S3). Of note, H2O2 was not fully consumed during the reactions, which levelled off after approximately 90–120 seconds. This is indicative of (partial) enzyme deactivation, likely due to incomplete active-site saturation. This suggests that soluble cello-oligosaccharides are not suitable for sustained LPMO turnover (see below, section 2.4).
The activity towards cellotetraose was further tested by applying the LsAA9A in the fast colourimetric assay using 2,6-DMP as a chromogenic reagent. This assay is based on the peroxidase side-reactivity of LPMOs, in which 2,6-DMP is oxidised to coerulignone in the presence of H2O2.27,28 This assay does not measure the substrate-dependent turnover of LPMOs; however, when carried out in the presence of the substrate cellotetraose, the DMP assay was effectively suppressed, indicating the interaction of the LsAA9A with cellotetraose41 (see Fig. S4). A similar effect was previously observed for the oxygen-reducing side reactivity of NcLPMO9C, an LPMO from Neurospora crassa, for cello-oligosaccharides with a higher degree of polymerisation.10
2.3 Post-reaction H2O2 accumulation indicates LPMO inactivation via self-oxidation
Fig. 4 presents the concentration of H2O2 measured over time following the completion of biocatalytic reactions using LsAA9A with varying concentrations of cellotetraose as substrate. A slow, but notable post-reaction accumulation of H2O2 levels was observed, suggesting that the enzyme undergoes inactivation.7,42–44 This is attributed to two primary pathways: (i) the reaction of reduced LPMO with molecular oxygen (O2), leading to the formation of H2O2, and (ii) a direct reaction of the reduced LPMO with H2O2 itself, a self-oxidative process that occurs in the absence of substrate.41 These pathways have been well-characterised for LPMOs acting on insoluble polysaccharides,41 where prolonged desorption from the substrate surface increases the risk of inactivation. Here, we show the same phenomenon for soluble cellotetraose. It should be noted that cello-oligosaccharides are probably not the preferred substrates, indicated by the high Michaelis–Menten constant (KM) of LsAA9A for cellotetraose of 0.22 mM (see section 2.4). Notably, the rates of H2O2 accumulation in these experiments inversely correlated with the initial substrate concentration. At higher cellotetraose concentrations, the enzyme appears to remain bound to the substrate for longer periods, thereby delaying desorption and the onset of inactivation. This extended substrate interaction offers a protective effect, reducing the frequency of uncoupled turnovers that generate damaging reactive oxygen species. These findings indicate that substrate availability plays a crucial role in modulating LPMO lifetime and activity. The self-inactivation of the enzyme after the reaction with cellotetraose was confirmed by determination of the post-reaction residual activity using the colourimetric 2,6-DMP assay (see Fig. S5). After the reaction, the LsAA9A retained only 11% of its initial activity, indicating substantial oxidative damage.2.4 LsAA9A kinetics with cellotetraose
To further understand the catalytic performance of LsAA9A, kinetic parameters were determined (Fig. 5) using the optical H2O2 sensor. Steady-state kinetics yielded a Michaelis–Menten constant (KM) of 0.22 ± 0.08 mM and a maximum reaction rate (Vmax) of 0.97 ± 0.64 μM s−1 (see Fig. S6 and S7). The LsAA9A showed substrate inhibition, with an apparent Ki of 212 nM, an effect that had already been previously observed for this enzyme and substrate.41 Specifically, the cleavage of fluorescently labelled cellotetraose revealed a KM of 43 ± 9 μM and a catalytic turnover rate (kcat) of 0.11 ± 0.01 s−1 at pH 7.0 and 37 °C, with reaction kinetics fitting well to classic Michaelis–Menten behaviour.45 However, these constants were obtained under different reaction conditions and, notably, in the absence of H2O2, making a direct comparison difficult. The obtained KM value of 43 ± 9 μM is low, compared to the one obtained in our study (0.22 mM). The difference could be partly due to the presence of aromatic groups at the reducing and non-reducing ends of the fluorescently labelled substrate cellotetraose.45 In support, the KM for another similar cello-oligosaccharide, cellopentaose, obtained for another known LPMO, NcAA9C, in the presence of H2O2, was also significantly higher (2.1 ± 0.3 mM, at 4 °C).163 Conclusions
In this study, an optical H2O2 sensor was validated for both H2O2-producing and -consuming redox enzymes in a homogenous reaction setup. Using glucose oxidase as a model enzyme, we show that the sensor enables reliable real-time detection of H2O2 formation, matching activity measurements obtained by spectrophotometric methods. More importantly, this study introduces a versatile and sensitive approach for monitoring LPMO activity based on H2O2 turnover. Importantly, these measurements are independent of the oxidation mode of the enzyme (C1, C4 or mixed oxidation) and can potentially be used with a variety of different substrates and with different reaction conditions. Despite the advantages, some limitations remain in the current implementation. Notably, there is a need to optimise the deoxygenation step preceding H2O2 detection. The currently required lag time between sample injection and flow cell detection introduces a delay that may affect the accurate timing of signal onset. While this does not affect steady-state comparisons, it may hinder the resolution of rapid kinetic events, especially in reactions with fast initial rates or short-lived intermediates. Overall, the optical sensor extends the portfolio of currently available methods for the determination of LPMO reactions and enables broader application for other peroxidases and peroxygenases.4 Experimental
4.1 Materials and reagents
Glucose oxidase from Aspergillus niger (type VII, lyophilized powder) was purchased from Merck. Cellotetraose (purity > 90%) was purchased from Megazyme. β-Glucosidase (10–50 units per mg) was purchased from Merck. D-Gluconic acid/D-glucono-δ-lactone assay kit was purchased from Megazyme.4.2 Production of LsAA9A
Production of LsAA9A from Lentinus similis was carried out as described previously.46 LsAA9A (EC 1.14.99.56, GenBank ALN96977.1), with a C-terminal double strep tag II, was ordered in the pET28a expression vector (ATG Biosynthetics, Germany) (Fig. S1). Chemically competent E. coli BL21 cells were transformed with the pET28a LsAA9A vector and plated onto LB agar containing 40 μg mL−1 kanamycin. A single colony was introduced into 4 mL LB medium (containing 40 μg mL−1 kanamycin) and cultured for 18 h. The culture was used to inoculate 400 mL of 2xYT auto induction medium supplemented with 100 μg mL−1 kanamycin and grown for 3.5 h at 37 °C, followed by a further incubation at 25 °C for 39 h (130 rpm). The cells were harvested by centrifugation (6000 rpm, 4 °C, 30 min) and stored at −20 °C until further use.The frozen cell pellets were thawed on ice for 1 h and resuspended in cold NP buffer (50 mM NaH2PO4, pH 8; 300 mM NaCl). The mixture was sonicated for 2 × 5 min at 50% power (Branson Sonifier, Sigma-Aldrich) and cell debris was removed by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, 1 h). The supernatant was filtered through a 0.2 μm syringe filter and loaded onto a 5 mL Strep-Tactin®X column (Cytiva) using an ÄKTA Pure chromatographic system (GE Healthcare). The column was washed with NP buffer to remove the unbound proteins. Finally, the apo-LsAA9A was eluted at once with NPB buffer (50 mM NaH2PO4, pH 8; 300 mM NaCl, 50 mM biotin). The enzyme solution was concentrated and rebuffered to potassium phosphate (50 mM, pH 6.0) using Vivaspin 20 centrifugal concentrators with a 10 kDa molecular weight cut-off. The protein concentration was determined by the Bradford method using bovine serum albumin for calibration.
000 rpm, 4 °C, 1 h). The supernatant was filtered through a 0.2 μm syringe filter and loaded onto a 5 mL Strep-Tactin®X column (Cytiva) using an ÄKTA Pure chromatographic system (GE Healthcare). The column was washed with NP buffer to remove the unbound proteins. Finally, the apo-LsAA9A was eluted at once with NPB buffer (50 mM NaH2PO4, pH 8; 300 mM NaCl, 50 mM biotin). The enzyme solution was concentrated and rebuffered to potassium phosphate (50 mM, pH 6.0) using Vivaspin 20 centrifugal concentrators with a 10 kDa molecular weight cut-off. The protein concentration was determined by the Bradford method using bovine serum albumin for calibration.
To generate copper-loaded LsAA9A, CuCl2 (10 mM in Milli-Q water) was slowly added to the apo-protein in a ratio apo-protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) copper of 1.5
copper of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, followed by incubation on ice for 2 h without any further desalting and concentration steps (from 6 × 300 mL cultures, approximately 18 mg of LsAA9A were produced).
1, followed by incubation on ice for 2 h without any further desalting and concentration steps (from 6 × 300 mL cultures, approximately 18 mg of LsAA9A were produced).
4.3 Glucose oxidase activity assay
The activity of the commercial glucose oxidase was assessed as described previously.30 In summary, the H2O2-dependent oxidation of the chromogenic reagent 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) by horseradish peroxidase (HRP) was measured spectrophotometrically, which depends on the formation of H2O2 by the glucose oxidase (GOD) (eqn (1) and (2)). The oxidation of ABTS (ε420 = 36 mM−1 cm−1) was continuously monitored by following the increase in absorbance at 420 nm.GOD activity was assayed with 10 mM glucose in 50 mM phosphate buffer, pH 6, using 7 U ml−1 HRP and 1 mM ABTS as chromogenic substrate. Reactions had a final volume of 200 μL and were measured in the 96-well plate format using a plate reader (BioTek Synergy, Agilent). Absorbance was continuously read at 570 nm for 25 min at 37 °C. One unit of enzyme activity was defined as the amount of GOD that oxidised 1 μmol of ABTS per minute under the specified assay conditions.
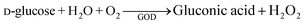 | (1) |
 | (2) |
4.4 Preparation of the H2O2 sensor
The H2O2 sensor used was first reported by Tjell et al.35 The setup used in these experiments (Fig. 1A and C) consists of luminescence-based sensing elements integrated into a flow cell. The sensing element contains a platinum-based catalyst and luminescent oxygen sensitive particles embedded in a host polymer. The catalyst converts H2O2 in the sample solution into molecular oxygen (O2), which is detected and quantified by the oxygen sensitive particles. To eliminate the background from dissolved O2 in the sample, the sample solution is pumped through sodium sulphite solution (Na2SO3, 2%), using a gas-permeable tube, to scavenge oxygen before entering the flow cell.The optical sensors were prepared from liquid sensor formulations as described previously.35 In brief, for O2 sensors, 2% palladium(II)-6,13,20,27-tetrakis(4-fluorophenyl) tetrabenzoporphyrin (PdTPTBPF) immobilized on PS/DVB beads dispersed in 7% Hydromed™ D7 in 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 EtOH/H2O were used. H2O2 sensors were prepared by adding 60 mg mL−1 of nanosilica-supported platinum nanoparticles (PtNP) to the O2 sensor formulation. The formulations were coated on a polyethylene glycol terephthalate (PET) support and glued into the flow-through. The cell was closed with pressure-sensitive adhesive tape (ThermalSeal RTS™, Excel Scientific, Inc., USA). The total internal volume of the sensor system (sample tube + deoxygenation tube + flow cell tube) was approx. 135 μL.
1 EtOH/H2O were used. H2O2 sensors were prepared by adding 60 mg mL−1 of nanosilica-supported platinum nanoparticles (PtNP) to the O2 sensor formulation. The formulations were coated on a polyethylene glycol terephthalate (PET) support and glued into the flow-through. The cell was closed with pressure-sensitive adhesive tape (ThermalSeal RTS™, Excel Scientific, Inc., USA). The total internal volume of the sensor system (sample tube + deoxygenation tube + flow cell tube) was approx. 135 μL.
For sensor read-outs a fibre-optic oxygen meter was used (FireSting, Pyroscience GmbH, Germany). A temperature sensor was placed in proximity to the flow cell to allow temperature-dependent O2 measurements. All data was recorded with the Pyro Oxygen Logger software (PyroScience GmbH, Germany) using two channels: channel 1 records the O2 formation by the platinum catalyst, while channel 2 records the baseline O2 level, which typically is below 2 mbar after deoxygenation. Channel 3 records the temperature.
The sensor was used for monitoring the activity of H2O2-generating and H2O2-consuming enzymes in solution. In all experiments, a sample flow of 50 μL min−1 was maintained using a syringe pump (New Era Instruments). Before each run, an extensive deoxygenation step was performed either overnight or for at least 8 hours by pumping degassed MQH2O through the sensor setup. The temperature was also recorded, and data points were logged every 3 seconds. Calibrations were performed with MQH2O and fresh stock solutions prepared by diluting 30% H2O2. Values of the calibration curve are averaged from continuous three-minute measurements (Fig. S2).
4.5 H2O2 sensor experiments for enzyme activity monitoring
GOD was assayed using the H2O2 sensor with 10 mM glucose in 50 mM phosphate buffer, pH 6, at 37 °C ± 1 °C. The reaction was performed using 2 mL conical tubes (Eppendorf, Germany). Reactions (excluding GOD) were prepared in a final volume of 1 mL, and the enzyme was added only after stabilisation of the H2O2 signal after approx. 10 min. All reactions were performed in a volume of 500 μL. The temperature was recorded, and data points were logged every 3 seconds. From the data points obtained by the sensor, concentrations of H2O2 were determined according to the calibration curve (Fig. S2).LsAA9A, as a H2O2-consuming enzyme, was measured with the same sensor setup. Ascorbic acid (AscA) was used as a reductant, and cellotetraose was supplied as substrate. One μM LsAA9A was assayed with soluble cellotetraose (0.05–1 mM), 50 μM H2O2 and 50 μM AscA in 50 mM phosphate buffer, pH 6, at 20 °C ± 1 °C. The reactions were performed in 2 mL conical tubes (Eppendorf). Reactions (excluding LsAA9A) were prepared in a final volume of 2 mL, and the enzyme was added only after stabilisation of the H2O2 signal after 20 min. The final reaction was performed in 1 mL volume. Control reactions that contained only cellotetraose at varying concentrations (0.1–1 mM), AscA (50 μM) in combination with H2O2 (50 μM), or cellotetraose (0.1 mM) in combination with H2O2 (50 μM) were performed to monitor the occurrence of non-enzymatic side reactions. LsAA9A was also studied in the absence and presence of cellotetraose. Additionally, the H2O2-consuming reaction by the selected LPMO was performed with and without reductant. A control using only Cu(II) was also performed as well as a reaction where the copper-dependent enzyme was chelated by EDTA 5 mM. The temperature was recorded, and data points were logged every 3 seconds. From the data points, given by the sensor, concentrations of H2O2 were determined according to the calibration curve (Fig. S2).
4.6 Activity of LsAA9A using a colorimetric assay
The H2O2-dependent activity of LsAA9A was assessed using 2,6-dimethoxyphenol (2,6-DMP) as described previously.27,28 The activity was measured before and after the reaction with cellotetraose (0.1 mM) and also in the presence and absence of cellotetraose (0.1–0.5 mM). Specifically, assays contained 10 mM 2,6-DMP, 50 μM H2O2 in 50 mM phosphate buffer, pH 6, and 4 μM of LsAA9A were used. Reactions had a total volume of 200 μL and were followed at 469 nm in a 96-well plate using a plate reader (BioTek Synergy, Agilent). One unit of LPMO activity was defined as the amount of LPMO that oxidises 2 μmol 2,6-DMP to 1 μmol coerulignone (ε469 = 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1) per min under the specified reaction conditions.
200 M−1 cm−1) per min under the specified reaction conditions.
4.7 Quantification of product formation by LsAA9A
The reaction with LsAA9A was performed by adding 1 μM LsAA9A to cellotetraose (0.1 mM), 50 μM H2O2 and 50 μM AscA in 50 mM phosphate buffer, pH 6, at 20 °C ± 1 °C. The reactions were performed using 2 mL conical tubes (Eppendorf) at a volume of 1 mL. 100 μL of the reaction mixture was sampled before the addition of the enzyme and after 20 s and 120 s. The collected samples were incubated with β-glucosidase at 50 °C overnight to release glucose and gluconic acid/gluconolactone from the LPMO reaction products. The resulting gluconic acid was quantified using the D-gluconic acid/D-glucono-δ-lactone assay kit from Megazyme. This assay detects gluconic acid or D-glucono-lactone by reacting it with gluconate kinase and gluconate-6-phosphate dehydrogenase, with the concomitant formation of NADPH that can be followed spectrophotometrically at 340 nm.4.8 Data processing
GraphPad Prism (10.4.1, Dotmatics) was used to generate graphics and modelling of the kinetics of LsAA9A. Activities were calculated from initial slopes of the reactions.Author contributions
D. K. and T. M. conceived the study. L. V. C. assisted in designing the experiments and conducted the experimental work. A. T. developed the prototype H2O2 optical sensor and established its setup, providing all materials. A. Ø. T. and T. M. contributed to the verification of H2O2 sensor and interpretation of the signal. L. V. C and D. K. were responsible for data interpretation and curation. M. D. F. R. supported the initial calibrations. L. V. C. and D. K. wrote the first draft of the manuscript. All authors contributed to the writing, review, and editing of the manuscript and approved the final version.Conflicts of interest
All authors declare that they have no conflicts of interest and no competing financial interests.Data availability
All kinetic measurements generated and analyzed during this study are fully presented in the main manuscript. Supporting data, including SDS-PAGE analyses confirming enzyme purity, calibration curves, and DNA sequences of the investigated enzymes, are provided in the supplementary information (SI) uploaded with the manuscript.Supplementary information is available. See DOI: https://doi.org/10.1039/d5re00332f.
Acknowledgements
This work was supported by the BioTechMed-Graz Young Researcher Group Program (D.K., L.V.C.). M.D.F.R. is grateful to the Ministry of Science, Innovation and Universities of Spain for a mobility grant “Salvador de Madariaga” (PRX22/00487).References
- D. Monti, G. Ottolina, G. Carrea and S. Riva, Chem. Rev., 2011, 111, 4111–4140 CrossRef CAS.
- A. J. C. Wahart, J. Staniland, G. J. Miller and S. C. Cosgrove, R. Soc. Open Sci., 2021, 8, 211572, DOI:10.1098/rsos.211572.
- M. Hobisch, D. Holtmann, P. Gomez de Santos, M. Alcalde, F. Hollmann and S. Kara, Biotechnol. Adv., 2021, 51, 107615 CrossRef CAS PubMed.
- B. Bissaro, A. Várnai, Å. K. Røhr and V. G. H. Eijsink, Microbiol. Mol. Biol. Rev., 2018, 82(4) DOI:10.1128/MMBR.00029-18.
- B. O. Burek, S. Bormann, F. Hollmann, J. Z. Bloh and D. Holtmann, Green Chem., 2019, 21, 3232–3249 RSC.
- G. Vaaje-Kolstad, B. Westereng, S. J. Horn, Z. Liu, H. Zhai, M. Sørlie and V. G. H. Eijsink, Science, 2010, 330, 219–222 CrossRef CAS PubMed.
- B. Bissaro, Å. K. Røhr, G. Müller, P. Chylenski, M. Skaugen, Z. Forsberg, S. J. Horn, G. Vaaje-Kolstad and V. G. H. Eijsink, Nat. Chem. Biol., 2017, 13(10), 1123–1128 CrossRef CAS.
- R. J. Quinlan, M. D. Sweeney, L. Lo Leggio, H. Otten, J.-C. N. Poulsen, K. S. Johansen, K. B. R. M. Krogh, C. I. Jørgensen, M. Tovborg, A. Anthonsen, T. Tryfona, C. P. Walter, P. Dupree, F. Xu, G. J. Davies and P. H. Walton, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15079–15084 CrossRef CAS.
- C. M. Phillips, W. T. Beeson, J. H. Cate and M. A. Marletta, ACS Chem. Biol., 2011, 6, 1399–1406 CrossRef CAS PubMed.
- T. Isaksen, B. Westereng, F. L. Aachmann, J. W. Agger, D. Kracher, R. Kittl, R. Ludwig, D. Haltrich, V. G. H. Eijsink and S. J. Horn, J. Biol. Chem., 2014, 289, 2632–2642 CrossRef CAS PubMed.
- K. S. Johansen, Biochem. Soc. Trans., 2016, 44, 143–149 CrossRef CAS.
- T. M. Vandhana, J.-L. Reyre, D. Sushmaa, J.-G. Berrin, B. Bissaro and J. Madhuprakash, New Phytol., 2022, 233, 2380–2396 CrossRef CAS PubMed.
- A. Munzone, V. G. H. Eijsink, J.-G. Berrin and B. Bissaro, Nat. Rev. Chem., 2024, 8, 106–119 CrossRef CAS PubMed.
- D. Kracher, T. Lanzmaier and L. V. Carneiro, Biochim. Biophys. Acta, Proteins Proteomics, 2024, 1872, 141012 CrossRef CAS PubMed.
- S. M. Jones, W. J. Transue, K. K. Meier, B. Kelemen and E. I. Solomon, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11916–11922 CrossRef CAS.
- L. Rieder, A. A. Stepnov, M. Sørlie and V. G. H. Eijsink, Biochemistry, 2021, 60, 3633–3643 CrossRef CAS PubMed.
- H. Chang, N. Gacias Amengual, A. Botz, L. Schwaiger, D. Kracher, S. Scheiblbrandner, F. Csarman and R. Ludwig, Nat. Commun., 2022, 13, 1–11 Search PubMed.
- R. Kont, B. Bissaro, V. G. H. Eijsink and P. Väljamäe, Nat. Commun., 2020, 11, 5786 CrossRef CAS PubMed.
- I. A. Christensen, V. G. H. Eijsink, A. A. Stepnov, G. Courtade and F. L. Aachmann, Biochemistry, 2023, 62, 1976–1993 CrossRef CAS PubMed.
- F. Filandr, D. Kavan, D. Kracher, C. V. F. P. Laurent, R. Ludwig, P. Man and P. Halada, Biomolecules, 2020, 10, 242 CrossRef CAS.
- A. Paradisi, E. M. Johnston, M. Tovborg, C. R. Nicoll, L. Ciano, A. Dowle, J. McMaster, Y. Hancock, G. J. Davies and P. H. Walton, J. Am. Chem. Soc., 2019, 141, 18585–18599 CrossRef CAS PubMed.
- G. R. Hemsworth, Essays Biochem., 2023, 67, 585–595 CrossRef PubMed.
- B. Westereng, J. S. M. Loose, G. Vaaje-Kolstad, F. L. Aachmann, M. Sørlie and V. G. H. Eijsink, Methods Mol. Biol., 2018, 1796, 219–246 CrossRef CAS PubMed.
- F. Calderaro, L. E. Bevers and M. A. van den Berg, Biomolecules, 2021, 11, 1098 CrossRef CAS.
- L. Schwaiger, A. Zenone, F. Csarman and R. Ludwig, Methods Enzymol., 2023, 679, 381–404 CAS.
- A. A. Stepnov and V. G. H. Eijsink, Methods Enzymol., 2023, 679, 163–189 CAS.
- E. Breslmayr, M. Hanžek, A. Hanrahan, C. Leitner, R. Kittl, B. Šantek, C. Oostenbrink and R. Ludwig, Biotechnol. Biofuels, 2018, 11, 1–13 CrossRef.
- E. Breslmayr, S. Daly, A. Požgajčić, H. Chang, T. Rezić, C. Oostenbrink and R. Ludwig, Biotechnol. Biofuels, 2019, 12(283) DOI:10.1186/S13068-019-1624-3.
- J. Ipsen, K. S. Johansen and S. Brander, Front. Microbiol., 2023, 14, 1128470 CrossRef.
- F. Filandr, P. Man, P. Halada, H. Chang, R. Ludwig and D. Kracher, Biotechnol. Biofuels, 2020, 13, 1–13 CrossRef.
- D. Wang, J. Li, A. C. Y. Wong, F. L. Aachmann and Y. S. Y. Hsieh, Biotechnol. Biofuels, 2018, 11, 1–11 CrossRef.
- M. B. Keller, C. Felby, C. A. Labate, V. O. A. Pellegrini, P. Higasi, R. K. Singh, I. Polikarpov and B. M. Blossom, Biotechnol. Lett., 2020, 42, 93–102 CrossRef CAS PubMed.
- J. Shen, P. T. Griffiths, S. J. Campbell, B. Utinger, M. Kalberer and S. E. Paulson, Sci. Rep., 2021, 11, 1–14 CrossRef.
- L. Schwaiger, F. Csarman, H. Chang, O. Golten, V. G. H. Eijsink and R. Ludwig, ACS Catal., 2024, 14, 1205–1219 CrossRef CAS.
- A. Tjell, B. Jud, R. Schaller-Ammann and T. Mayr, Sens. Actuators, B, 2024, 400, 134904 CrossRef CAS.
- A. I. Tjell, L. E. Meyer, B. Jud, S. Kara and T. Mayr, React. Chem. Eng., 2024, 9, 777–781 RSC.
- R. Tokin, K. E. H. Frandsen, J. Ø. Ipsen, L. Lo Leggio, M. M. Poojary, J. G. Berrin, S. Grisel, S. Brander, P. E. Jensen and K. S. Johansen, New Phytol., 2021, 232, 1337–1349 CrossRef CAS PubMed.
- T. Tandrup, T. Tryfona, K. E. H. Frandsen, K. S. Johansen, P. Dupree and L. Lo Leggio, Biochemistry, 2020, 59, 3347–3358 CrossRef CAS.
- T. Tandrup, L. Lo Leggio, F. Meilleur and M. J. Romao, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2023, 79, 1–7 CrossRef CAS.
- M. Frommhagen, A. H. Westphal, W. J. H. van Berkel and M. A. Kabel, Front. Microbiol., 2018, 9, 358610 Search PubMed.
- S. Brander, R. Tokin, J. Ipsen, P. E. Jensen, C. Hernández-Rollán, M. H. H. Nørholm, L. Lo Leggio, P. Dupree and K. S. Johansen, ACS Catal., 2021, 11, 13848–13859 CrossRef CAS.
- S. Kuusk, V. G. H. Eijsink and P. Väljamäe, J. Biol. Chem., 2023, 299, 105094 CrossRef CAS.
- G. Müller, P. Chylenski, B. Bissaro, V. G. H. Eijsink and S. J. Horn, Biotechnol. Biofuels, 2018, 11, 1–17 CrossRef.
- D. Kracher, M. Andlar, P. G. Furtmüller and R. Ludwig, J. Biol. Chem., 2017, 293, 1676 CrossRef.
- K. E. H. Frandsen, T. J. Simmons, P. Dupree, J. C. N. Poulsen, G. R. Hemsworth, L. Ciano, E. M. Johnston, M. Tovborg, K. S. Johansen, P. Von Freiesleben, L. Marmuse, S. Fort, S. Cottaz, H. Driguez, B. Henrissat, N. Lenfant, F. Tuna, A. Baldansuren, G. J. Davies, L. Lo Leggio and P. H. Walton, Nat. Chem. Biol., 2016, 12(4), 298–303 CrossRef CAS PubMed.
- J. Zhao, Y. Zhuo, D. E. Diaz, M. Shanmugam, A. J. Telfer, P. J. Lindley, D. Kracher, T. Hayashi, L. S. Seibt, F. J. Hardy, O. Manners, T. M. Hedison, K. A. Hollywood, R. Spiess, K. M. Cain, S. Diaz-Moreno, N. S. Scrutton, M. Tovborg, P. H. Walton, D. J. Heyes and A. P. Green, J. Am. Chem. Soc., 2023, 145, 20672–20682 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |