Open Access Article
Open Access Article[BMIM]OAc promoted one-pot synthesis of pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-5-ones and their antimicrobial activity
Savan S. Bhalodiya a,
Mehul P. Parmar
a,
Mehul P. Parmar a,
Shana Balachandran
a,
Shana Balachandran b,
Chirag D. Patel
b,
Chirag D. Patel a,
Arijit Nandi
a,
Arijit Nandi cd,
Anwesha Das
cd,
Anwesha Das ce,
Madan Kumar Arumugam
ce,
Madan Kumar Arumugam b and
Hitendra M. Patel
b and
Hitendra M. Patel *a
*a
aDepartment of Chemistry, Sardar Patel University, Vallabh Vidyanagar, Near University Circle, Vallabh Vidyanagar-388120, Gujarat, India. E-mail: hm_patel@spuvvn.edu; sa1bhalodiya@gmail.com; mpparmar1997@gmail.com; chiragkumarp23@gmail.com
bCancer Biology Lab, Center for Molecular and Nanomedical Sciences, Sathyabama Institute of Science and Technology, Chennai-600119, Tamil Nadu, India. E-mail: shanabalachandran@gmail.com; madankumar@sathyabama.ac.in; madankumarbio@gmail.com
cDepartment of Pharmacy, Sanaka Educational Trust Group of Institutions (SETGOI), Durgapur, West Bengal 713212, India. E-mail: arijitnandi57@gmail.com; dasanwesha.096@gmail.com
dInstitute for Molecular Bioscience, The University of Queensland, Brisbane, 4072, Australia
eSchool of Pharmacy and Pharmaceutical Sciences, The University of Queensland, Brisbane, Queensland 4072 Queensland, Australia
First published on 2nd January 2026
Abstract
A one-pot method was developed for synthesizing pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-5-ones using the recyclable ionic liquid catalyst [BMIM]OAc in ethanol under reflux conditions. This approach achieved high yields (76–95%) across various substrates along with straightforward product isolation and catalyst recyclability. The synthesized compounds 4(a–l) were tested for antimicrobial activity against Gram-positive and Gram-negative bacteria, and fungal strains. Notably, compounds 4c and 4e showed strong antibacterial effects against S. aureus (MIC: 8 µg mL−1), while 4a, 4b, and 4f effectively inhibited E. coli (MIC: 8 µg mL−1). Compound 4l exhibited notable antifungal activity against C. albicans (MIC: 8 µg mL−1), outperforming the standard drugs. Cytotoxicity assessment on HEK293 cells revealed low toxicity in most derivatives (IC50 > 50 µM), with halogenated analogs (4c, 4e) displaying moderate activity but remaining less toxic than doxorubicin. Molecular docking and dynamics simulations confirmed stable interactions of key compounds (4c, 4e, 4f) with bacterial Tet repressor class D and fungal sterol 14-α demethylase, consistent with experimental data. Overall, this ionic liquid-catalyzed approach provides an efficient route to bioactive heterocycles with promising antimicrobial and anticancer potential.
1 Introduction
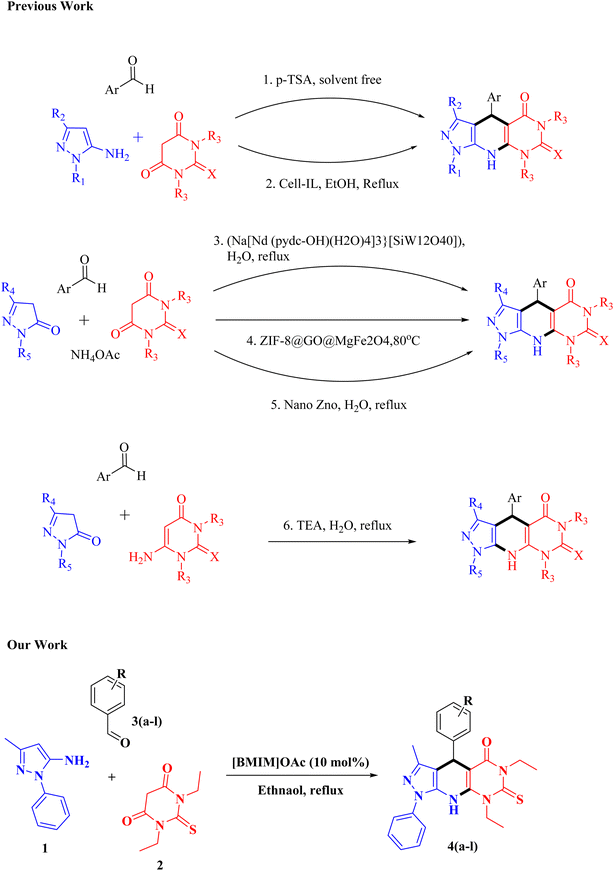
Heterocyclic structures are crucial in drug discovery, appearing in over 70% of small-molecule drugs approved in recent decades due to their ability to finely adjust properties like lipophilicity, polarity, hydrogen bonding, and ADME profiles.1–4 Synthetic advances have enabled access to a wide variety of these compounds, speeding up medicinal chemistry efforts.5 Fused heterocycles, for example, pyrazolo-pyrimidines, are highly regarded as privileged scaffolds because their rigid, nucleobase-like forms enhance selective binding to ATP-binding enzymes.6–8 Multicomponent reactions (MCRs) are highly valuable in modern synthetic chemistry due to their efficiency, sustainability, and versatility in constructing complex molecules in a single step.9–13 These reactions adhere to green chemistry principles by often removing the necessity for intermediate purification and minimizing reagent consumption.14–17 MCRs have broad applications across pharmaceuticals, materials science, and agrochemicals, serving as a robust platform to generate extensive libraries of bioactive compounds, functional materials, and innovative molecular architectures critical for both research progress and industrial use.18–20Recently, ionic liquids have become prominent catalysts in the synthesis of heterocyclic structures due to their dual role as green solvents and catalysts.21,22 Ionic liquids act green and sustainable catalyst and solvent, the cation and anion combination can be changed for specific reaction conditions, enhances reaction efficiency.23 Numerous one-pot MCRs conducted in ionic liquids have been reported for synthesizing diverse heterocycles such as pyrimidine, quinoline, quinazoline, pyrimido-quinoline, and pyrido-pyrimidine derivatives.24–27 Pyrazolo, pyridine, and pyrimidine scaffolds possess various biological activity because of producing nitrogen-rich, planar frameworks capable of hydrogen bonding and π–π interactions at biomolecular sites.28–30 Owing to these features, they are considered privileged scaffolds in medicinal chemistry and have yielded diverse bioactive compounds, into clinical studies.6,31,32 These heterocycles exhibit antimicrobial, antifungal, anti-inflammatory, and anticancer activities supported by SAR studies.33–37 Such versatility underscores their potential for further development as antimicrobial agents. Some of the marketed drugs bearing pyrazole, pyridine, and pyrimidine scaffold were shown in Fig. 1.
According to literature, the rational design of the target molecules was guided by established SAR trends of barbituric and thiobarbituric derivatives. Replacement of the carbonyl oxygen with sulphur at C-2 increases lipophilicity and polarizability, enhancing passive membrane diffusion and soft–soft interactions with biological targets compared to the parent oxygen analogues.38 Published studies report superior antibacterial profiles for thiobarbiturates versus barbiturates, attributed to altered hydrogen-bonding geometry and improved intracellular accumulation. Additionally, N-alkyl substitution modulates lipophilicity and steric bulk; ethyl chains provide an optimal balance between hydrophobicity and solubility, improving bacterial membrane penetration relative to methyl substituents. Therefore, systematic incorporation of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S group and variation of N-alkyl chain length (methyl vs. ethyl) was selected to evaluate how physicochemical changes translate into antimicrobial potency.39,40
S group and variation of N-alkyl chain length (methyl vs. ethyl) was selected to evaluate how physicochemical changes translate into antimicrobial potency.39,40
Several protocols exist for constructing pyrazolo-pyrido-pyrimidine scaffolds by employing various catalyst like p-TSA,41 cellulose supported acidic ionic liquid,42 (Na{[Nd (pydc-OH)(H2O)4]3}[SiW12O40]),43 ZIF-8@GO@MgFe2O4,44 Nano-ZnO,45 triethyl amine,46 conditions. To the best of our knowledge, there is a lack of alternative methodologies for synthesizing pyrazolo-pyrido-pyrimidine scaffolds has been reported in the current literature. Therefore, exploring new and diverse strategies for their synthesis remains essential due to their significant importance.
To the best of our knowledge, this is the first report describing a recyclable ionic-liquid-promoted one-pot MCR synthesis of pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-5-ones. Compared with previously published protocols employing different catalysts, the present methodology offers several distinctive advantages, including milder reaction conditions, significantly higher yields, short reaction time, simple product isolation, and efficient catalyst recyclability. Furthermore, the current work not only provides an environmentally sustainable synthetic route but also delivers biologically active compounds with promising antimicrobial profiles, thereby demonstrating practical medicinal relevance (Scheme 1).
2 Experimental
2.1 Synthetic protocol for pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-5-ones 4(a–l)
Using 10 mol% [BMIM]OAc as the catalyst, compounds 4(a–l) were synthesized by reacting 5-amino-3-methyl-1-phenylpyrazole (1, 1.0 mmol), 1,3-diethylthiobarbituric acid (2, 1.0 mmol), and the corresponding aromatic aldehydes (3(a–l), 1.0 mmol) in ethanol under reflux conditions. The progress of the reaction was monitored by thin-layer chromatography (TLC) using silica gel 60 F254 plates with an ethyl acetate/hexane (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, v/v) solvent system. After reaction completion, the mixture was allowed to cool to room temperature, during which a solid product precipitated in the reaction flask. The reaction mixture was then quenched by the addition of water. The resulting solid was collected by filtration and washed thoroughly with small amounts of ethanol followed by hot water to afford the pure product.
7, v/v) solvent system. After reaction completion, the mixture was allowed to cool to room temperature, during which a solid product precipitated in the reaction flask. The reaction mixture was then quenched by the addition of water. The resulting solid was collected by filtration and washed thoroughly with small amounts of ethanol followed by hot water to afford the pure product.
2.2 Analytical data of synthesized compounds 4(a–l)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1137 (s, C
O), 1137 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.42 (s, 1H, N–H), 7.32 (d, 2H, J = 4 Hz, Ar–H), 7.08 (d, 2H, J = 8 Hz, Ar–H), 6.95 (s, 1H, Ar–H), 6.62 (t, 2H, J = 4 Hz, Ar–H), 6.53 (d, 2H, J = 4 Hz, Ar–H), 5.37 (s, 1H, C–H), 4.41 (q, 4H, J = 8 Hz, CH2), 2.07 (s, 3H, CH3), 1.30 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 171.97, 162.81, 160.82, 156.82, 152.44, 151.08, 144.83, 132.54, 131.41, 125.08, 124.24, 121.15, 115.34, 133.37, 84.89, 47.28, 43.63, 17.19, 13.11, 12.04. Molecular weight: 461.95, MS(ESI m/z): 462.82.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.42 (s, 1H, N–H), 7.32 (d, 2H, J = 4 Hz, Ar–H), 7.08 (d, 2H, J = 8 Hz, Ar–H), 6.95 (s, 1H, Ar–H), 6.62 (t, 2H, J = 4 Hz, Ar–H), 6.53 (d, 2H, J = 4 Hz, Ar–H), 5.37 (s, 1H, C–H), 4.41 (q, 4H, J = 8 Hz, CH2), 2.07 (s, 3H, CH3), 1.30 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 171.97, 162.81, 160.82, 156.82, 152.44, 151.08, 144.83, 132.54, 131.41, 125.08, 124.24, 121.15, 115.34, 133.37, 84.89, 47.28, 43.63, 17.19, 13.11, 12.04. Molecular weight: 461.95, MS(ESI m/z): 462.82.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1134 (s, C
O), 1134 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.86 (s, 1H, N–H), 7.62 (t, 1H, J = 8 Hz, Ar–H), 7.51 (d, 2H, J = 8 Hz, Ar–H), 7.45 (d, 2H, J = 4 Hz, Ar–H), 7.36 (d, 2H, J = 8 Hz, Ar–H), 7.18 (d, 2H, J = 8 Hz, Ar–H), 5.51 (s, 1H, C–H), 4.42 (q, 4H, J = 8 Hz, CH2), 2.12 (s, 3H, CH3), 1.46 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 174.36, 167.85, 161.17, 145.70, 142.78, 140.57, 138.11, 135.76, 133.37, 129.99, 129.44, 128.61., 127.54, 125.60, 85.90, 47.31, 43.92, 43.07. Molecular weight: 506.40, MS(ESI m/z): 507.37.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.86 (s, 1H, N–H), 7.62 (t, 1H, J = 8 Hz, Ar–H), 7.51 (d, 2H, J = 8 Hz, Ar–H), 7.45 (d, 2H, J = 4 Hz, Ar–H), 7.36 (d, 2H, J = 8 Hz, Ar–H), 7.18 (d, 2H, J = 8 Hz, Ar–H), 5.51 (s, 1H, C–H), 4.42 (q, 4H, J = 8 Hz, CH2), 2.12 (s, 3H, CH3), 1.46 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 174.36, 167.85, 161.17, 145.70, 142.78, 140.57, 138.11, 135.76, 133.37, 129.99, 129.44, 128.61., 127.54, 125.60, 85.90, 47.31, 43.92, 43.07. Molecular weight: 506.40, MS(ESI m/z): 507.37.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1121 (s, C
O), 1121 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.89 (s, 1H, N–H), 7.61 (t, 2H, J = 8 Hz, Ar–H), 7.52 (d, 2H, J = 8 Hz, Ar–H), 7.46 (q, 2H, J = 8 Hz, Ar–H), 7.36 (d, 2H, J = 8 Hz, Ar–H), 7.11 (s, 1H, Ar–H), 5.56 (s, 1H, C–H), 4.40 (q, 4H, J = 8 Hz, CH2), 2.10 (s, 3H, CH3), 1.43 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 171.51, 163.13, 160.09, 140.27, 139.00, 136.86, 134.45, 132.11, 130.06, 129.70, 127.03, 122.42, 114.03, 112.99, 82.23, 48.42, 42.68, 42.13, 17.84, 12.25, 11.58. Molecular weight: 445.50, MS(ESI m/z): 444.83.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.89 (s, 1H, N–H), 7.61 (t, 2H, J = 8 Hz, Ar–H), 7.52 (d, 2H, J = 8 Hz, Ar–H), 7.46 (q, 2H, J = 8 Hz, Ar–H), 7.36 (d, 2H, J = 8 Hz, Ar–H), 7.11 (s, 1H, Ar–H), 5.56 (s, 1H, C–H), 4.40 (q, 4H, J = 8 Hz, CH2), 2.10 (s, 3H, CH3), 1.43 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 171.51, 163.13, 160.09, 140.27, 139.00, 136.86, 134.45, 132.11, 130.06, 129.70, 127.03, 122.42, 114.03, 112.99, 82.23, 48.42, 42.68, 42.13, 17.84, 12.25, 11.58. Molecular weight: 445.50, MS(ESI m/z): 444.83.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1532 (s, NO2), 1364 (s, NO2), 1127 (s, C
O), 1532 (s, NO2), 1364 (s, NO2), 1127 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.94 (s, 1H, N–H), 7.63 (t, 2H, J = 8 Hz, Ar–H), 7.48 (m, 3H, Ar–H), 7.38 (d, 2H, J = 4 Hz, Ar–H), 3.32 (t, 2H, J = 8 Hz, Ar–H), 5.66 (s, 1H, C–H), 4.45 (q, 4H, J = 8 Hz, CH2), 2.06 (s, 3H, CH3), 1.35 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.44, 167.21, 164.63, 144.57, 143.24, 141.77, 139.98, 138.55, 136.11, 133.96, 131.72, 130.00, 127.87, 116.93, 83.37, 48.11, 42.90, 42.15, 18.05, 12.68, 11.80. Molecular weight: 472.51, MS(ESI m/z): 471.29.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.94 (s, 1H, N–H), 7.63 (t, 2H, J = 8 Hz, Ar–H), 7.48 (m, 3H, Ar–H), 7.38 (d, 2H, J = 4 Hz, Ar–H), 3.32 (t, 2H, J = 8 Hz, Ar–H), 5.66 (s, 1H, C–H), 4.45 (q, 4H, J = 8 Hz, CH2), 2.06 (s, 3H, CH3), 1.35 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.44, 167.21, 164.63, 144.57, 143.24, 141.77, 139.98, 138.55, 136.11, 133.96, 131.72, 130.00, 127.87, 116.93, 83.37, 48.11, 42.90, 42.15, 18.05, 12.68, 11.80. Molecular weight: 472.51, MS(ESI m/z): 471.29.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1113 (s, C
O), 1113 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.66 (s, 1H, N–H), 7.57 (s, 1H, Ar–H), 7.55 (s, 1H, Ar–H), 7.53 (d, 5H, J = 4 Hz, Ar–H), 7.40 (t, 2H, J = 4 Hz, Ar–H), 5.38 (s, 1H, C–H), 4.43 (q, 4H, J = 8 Hz, CH2), 2.14 (s, 3H, CH3), 1.40 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 171.99, 165.88, 163.23, 145.43, 143.36, 141.09, 139.04, 135.48, 133.49, 131.91, 129.03, 124.81, 122.64, 120.02, 117.15, 112.48, 84.36, 49.47, 43.74, 42.72, 18.35, 13.06, 12.18. Molecular weight: 461.95, MS(ESI m/z): 460.38.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.66 (s, 1H, N–H), 7.57 (s, 1H, Ar–H), 7.55 (s, 1H, Ar–H), 7.53 (d, 5H, J = 4 Hz, Ar–H), 7.40 (t, 2H, J = 4 Hz, Ar–H), 5.38 (s, 1H, C–H), 4.43 (q, 4H, J = 8 Hz, CH2), 2.14 (s, 3H, CH3), 1.40 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 171.99, 165.88, 163.23, 145.43, 143.36, 141.09, 139.04, 135.48, 133.49, 131.91, 129.03, 124.81, 122.64, 120.02, 117.15, 112.48, 84.36, 49.47, 43.74, 42.72, 18.35, 13.06, 12.18. Molecular weight: 461.95, MS(ESI m/z): 460.38.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1129 (s, C
O), 1129 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.44 (s, 1H, N–H), 7.34 (d, 2H, J = 8 Hz, Ar–H), 7.12 (t, 3H, J = 8 Hz, Ar–H), 6.88 (d, 2H, J = 4 Hz, Ar–H), 6.57 (d, 2H, J = 4 Hz, Ar–H), 5.41 (s, 1H, C–H), 4.40 (q, 4H, J = 8 Hz, CH2), 2.10 (s, 3H, CH3), 1.37 (t, 6H, J = 8 Hz, CH3); 13C NMR (126 MHz, DMSO-d6) δ: 173.73, 167.90, 165.84, 144.05, 141.82, 139.53, 139.16, 135.50, 133.21, 131.04, 129.11, 127.35, 122.82, 122.02, 119.80, 117.25, 83.84, 47.80, 43.21, 42.59, 18.93, 12.74, 11.77. Molecular weight: 506.40, MS(ESI m/z): 507.57.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.44 (s, 1H, N–H), 7.34 (d, 2H, J = 8 Hz, Ar–H), 7.12 (t, 3H, J = 8 Hz, Ar–H), 6.88 (d, 2H, J = 4 Hz, Ar–H), 6.57 (d, 2H, J = 4 Hz, Ar–H), 5.41 (s, 1H, C–H), 4.40 (q, 4H, J = 8 Hz, CH2), 2.10 (s, 3H, CH3), 1.37 (t, 6H, J = 8 Hz, CH3); 13C NMR (126 MHz, DMSO-d6) δ: 173.73, 167.90, 165.84, 144.05, 141.82, 139.53, 139.16, 135.50, 133.21, 131.04, 129.11, 127.35, 122.82, 122.02, 119.80, 117.25, 83.84, 47.80, 43.21, 42.59, 18.93, 12.74, 11.77. Molecular weight: 506.40, MS(ESI m/z): 507.57.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1136 (s, C
O), 1136 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.51 (s, 1H, N–H), 8.20 (d, 3H, J = 4 Hz, Ar–H), 8.07 (t, 2H, J = 2 Hz, Ar–H), 7.88 (t, 2H, J = 4 Hz, Ar–H), 6.63 (q, 2H, J = 4 Hz, Ar–H), 5.50 (s, 1H, C–H), 4.49 (q, 4H, J = 8 Hz, CH2), 2.05 (s, 3H, CH3), 1.30 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.45, 167.16, 165.79, 144.47, 142.57, 139.21, 137.30, 135.17, 131.50, 128.27, 123.63, 121.23, 119.07, 116.59, 114.43, 114.28, 83.22, 43.30, 42.29, 18.32, 12.58, 11.67. Molecular weight: 445.50, MS(ESI m/z): 444.92.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.51 (s, 1H, N–H), 8.20 (d, 3H, J = 4 Hz, Ar–H), 8.07 (t, 2H, J = 2 Hz, Ar–H), 7.88 (t, 2H, J = 4 Hz, Ar–H), 6.63 (q, 2H, J = 4 Hz, Ar–H), 5.50 (s, 1H, C–H), 4.49 (q, 4H, J = 8 Hz, CH2), 2.05 (s, 3H, CH3), 1.30 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.45, 167.16, 165.79, 144.47, 142.57, 139.21, 137.30, 135.17, 131.50, 128.27, 123.63, 121.23, 119.07, 116.59, 114.43, 114.28, 83.22, 43.30, 42.29, 18.32, 12.58, 11.67. Molecular weight: 445.50, MS(ESI m/z): 444.92.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1521 (s, NO2), 1352 (s, NO2), 1135 (s, C
O), 1521 (s, NO2), 1352 (s, NO2), 1135 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.52 (s, 1H, N–H), 7.64 (d, 3H, J = 4 Hz, Ar–H), 7.59 (d, 1H, J = 4 Hz, Ar–H), 7.26 (d, 3H, J = 8 Hz, Ar–H), 7.12 (t, 2H, J = 8 Hz, Ar–H), 5.55 (s, 1H, C–H), 4.37 (q, 4H, J = 8 Hz, CH2), 2.09 (s, 3H, CH3), 1.13 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 174.69, 163.38, 159.76, 144.57, 137.87, 135.78, 131.68, 130.26, 130.02, 128.70, 128.18, 126.49, 125.31, 123.50, 122.70, 117.14, 116.65, 83.84, 48.46, 43.20, 42.38, 18.53, 12.81, 12.13. Molecular weight: 472.51, MS(ESI m/z): 471.08.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.52 (s, 1H, N–H), 7.64 (d, 3H, J = 4 Hz, Ar–H), 7.59 (d, 1H, J = 4 Hz, Ar–H), 7.26 (d, 3H, J = 8 Hz, Ar–H), 7.12 (t, 2H, J = 8 Hz, Ar–H), 5.55 (s, 1H, C–H), 4.37 (q, 4H, J = 8 Hz, CH2), 2.09 (s, 3H, CH3), 1.13 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 174.69, 163.38, 159.76, 144.57, 137.87, 135.78, 131.68, 130.26, 130.02, 128.70, 128.18, 126.49, 125.31, 123.50, 122.70, 117.14, 116.65, 83.84, 48.46, 43.20, 42.38, 18.53, 12.81, 12.13. Molecular weight: 472.51, MS(ESI m/z): 471.08.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1127 (s, C
O), 1127 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.63 (s, 1H, N–H), 7.44 (d, 3H, J = 4 Hz, Ar–H), 7.39 (q, 1H, J = 4 Hz, Ar–H), 7.07 (d, 2H, J = 8Hz, Ar–H), 7.79 (d, 2H, J = 8 Hz, Ar–H), 6.53 (q, 1H, J = 4 Hz, Ar–H), 5.35 (s, 1H, C–H), 4.46 (q, 4H, J = 8 Hz, CH2), 2.08 (s, 3H, CH3), 1.33 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 174.73, 165.43, 161.45, 144.89, 142.11, 137.22, 135.59, 133.88, 131.72, 128.79, 125.85, 123.76, 121.64, 120.27, 116.63, 115.18, 83.45, 48.81, 43.36, 42.49, 18.09, 12.64, 11.89. Molecular weight: 461.95, MS(ESI m/z): 460.92.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.63 (s, 1H, N–H), 7.44 (d, 3H, J = 4 Hz, Ar–H), 7.39 (q, 1H, J = 4 Hz, Ar–H), 7.07 (d, 2H, J = 8Hz, Ar–H), 7.79 (d, 2H, J = 8 Hz, Ar–H), 6.53 (q, 1H, J = 4 Hz, Ar–H), 5.35 (s, 1H, C–H), 4.46 (q, 4H, J = 8 Hz, CH2), 2.08 (s, 3H, CH3), 1.33 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 174.73, 165.43, 161.45, 144.89, 142.11, 137.22, 135.59, 133.88, 131.72, 128.79, 125.85, 123.76, 121.64, 120.27, 116.63, 115.18, 83.45, 48.81, 43.36, 42.49, 18.09, 12.64, 11.89. Molecular weight: 461.95, MS(ESI m/z): 460.92.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1131 (s, C
O), 1131 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (400 MHz, DMSO-d6) δ: 9.74 (s, 1H, N–H), 7.43 (d, 1H, J = 8 Hz, Ar–H), 7.39 (s, 1H, Ar–H), 7.16 (q, 2H, J = 4 Hz, Ar–H), 7.10 (d, 2H, J = 8 Hz, Ar–H), 6.89 (d, 2H, J = 8 Hz, Ar–H), 6.80 (s, 1H, Ar–H), 5.34 (s, 1H, C–H), 4.41 (q, 4H, J = 8 Hz, CH2), 2.05 (s, 3H, CH3), 1.32 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.87, 164.47, 162.16, 144.52, 143.08, 139.83, 137.21, 134.47, 130.55, 129.65, 128.86, 125.68, 123.58, 121.59, 118.51, 116.67, 84.33, 49.02, 43.56, 42.93, 18.81, 12.80, 11.95. Molecular weight: 506.40, MS(ESI m/z): 507.62.
S) 1H NMR (400 MHz, DMSO-d6) δ: 9.74 (s, 1H, N–H), 7.43 (d, 1H, J = 8 Hz, Ar–H), 7.39 (s, 1H, Ar–H), 7.16 (q, 2H, J = 4 Hz, Ar–H), 7.10 (d, 2H, J = 8 Hz, Ar–H), 6.89 (d, 2H, J = 8 Hz, Ar–H), 6.80 (s, 1H, Ar–H), 5.34 (s, 1H, C–H), 4.41 (q, 4H, J = 8 Hz, CH2), 2.05 (s, 3H, CH3), 1.32 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.87, 164.47, 162.16, 144.52, 143.08, 139.83, 137.21, 134.47, 130.55, 129.65, 128.86, 125.68, 123.58, 121.59, 118.51, 116.67, 84.33, 49.02, 43.56, 42.93, 18.81, 12.80, 11.95. Molecular weight: 506.40, MS(ESI m/z): 507.62.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1129 (s, C
O), 1129 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S); 1H NMR (500 MHz, DMSO-d6) δ: 9.82 (s, 1H, N–H), 7.45 (d, 2H, J = 8 Hz, Ar–H), 7.26 (d, 2H, J = 8 Hz, Ar–H), 7.11 (m, 4H, Ar–H), 6.87 (d, 1H, J = 4Hz, Ar–H), 5.47 (s, 1H, C–H), 4.39 (q, 4H, J = 8 Hz, CH2), 2.08 (s, 3H, CH3), 1.31 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.06, 163.93, 161.61, 143.60, 142.11, 136.99, 135.31, 133.44, 131.46, 128.41, 123.55, 122.61, 117.37, 116.73, 83.80, 49.23, 43.46, 42.63, 18.89, 12.38, 11.35. Molecular weight: 495.51, MS(ESI m/z): 496.56.
S); 1H NMR (500 MHz, DMSO-d6) δ: 9.82 (s, 1H, N–H), 7.45 (d, 2H, J = 8 Hz, Ar–H), 7.26 (d, 2H, J = 8 Hz, Ar–H), 7.11 (m, 4H, Ar–H), 6.87 (d, 1H, J = 4Hz, Ar–H), 5.47 (s, 1H, C–H), 4.39 (q, 4H, J = 8 Hz, CH2), 2.08 (s, 3H, CH3), 1.31 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 173.06, 163.93, 161.61, 143.60, 142.11, 136.99, 135.31, 133.44, 131.46, 128.41, 123.55, 122.61, 117.37, 116.73, 83.80, 49.23, 43.46, 42.63, 18.89, 12.38, 11.35. Molecular weight: 495.51, MS(ESI m/z): 496.56.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1131 (s, C
O), 1131 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 1H NMR (500 MHz, DMSO-d6) δ: 9.89 (s, 1H, N–H), 7.61 (t, 2H, J = 8 Hz, Ar–H), 7.52 (d, 2H, J = 8 Hz, Ar–H), 7.46 (q, 2H, J = 8 Hz, Ar–H), 7.36 (d, 2H, J = 8 Hz, Ar–H), 7.11 (s, 1H, Ar–H), 5.56 (s, 1H, C–H), 4.42 (q, 4H, J = 8 Hz, CH2), 2.10 (s, 3H, CH3), 1.43 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 175.04, 167.36, 165.50, 143.71, 143.28, 137.62, 132.56, 131.65, 131.59, 128.35, 123.61, 122.79, 117.37, 116.78, 85.35, 50.17, 44.60, 43.37, 19.20, 14.25, 13.38. Molecular weight: 452.52, MS(ESI m/z): 453.570.
S) 1H NMR (500 MHz, DMSO-d6) δ: 9.89 (s, 1H, N–H), 7.61 (t, 2H, J = 8 Hz, Ar–H), 7.52 (d, 2H, J = 8 Hz, Ar–H), 7.46 (q, 2H, J = 8 Hz, Ar–H), 7.36 (d, 2H, J = 8 Hz, Ar–H), 7.11 (s, 1H, Ar–H), 5.56 (s, 1H, C–H), 4.42 (q, 4H, J = 8 Hz, CH2), 2.10 (s, 3H, CH3), 1.43 (t, 6H, J = 8 Hz, CH3) 13C NMR (126 MHz, DMSO-d6) δ: 175.04, 167.36, 165.50, 143.71, 143.28, 137.62, 132.56, 131.65, 131.59, 128.35, 123.61, 122.79, 117.37, 116.78, 85.35, 50.17, 44.60, 43.37, 19.20, 14.25, 13.38. Molecular weight: 452.52, MS(ESI m/z): 453.570.2.3 Biological screening
2.4 Molecular docking
The crystal structures of the Tet repressor class D bound to tetracycline (PDB ID: 2VKE) and the sterol 14-alpha demethylase complexed with the tetrazole-based antifungal candidate VT1161 (PDB ID: 5TZ1) were obtained from the RCSB Protein Data Bank. Both protein structures were processed using the Protein Preparation Wizard in Schrödinger (version 2017_2) 47, 48 to eliminate water molecules, reconstruct missing loops and side chains, and add hydrogen atoms for proper structural optimization. Protonation states and partial charges were assigned using the OPLS3 force field. The protein structures were subsequently subjected to restrained minimization until the root-mean-square deviation (RMSD) of non-hydrogen atoms reached 0.3 Å. Ligand preparation was carried out with the LigPrep module in the Schrödinger 2017-2 suite to generate possible tautomers and predict ionization states at pH 7.0 ± 2.0 using Epik. Receptor grids were then constructed around the co-crystallized ligand binding site of each protein's crystal structure. Finally, molecular docking was performed using the Glide XP protocol with the optimized receptor models.2.5 MD simulations
MD simulations of the docked protein–ligand complexes were performed using the Desmond module within the Schrödinger software suite. The simulation system was constructed in an orthorhombic box via the System Builder panel, employing the simple point charge (SPC) model to represent water molecules. Simulations were carried out under an NPT ensemble at 310 K and 1.013 bar for a duration of 100 ns. Results from the molecular dynamics analyses were evaluated using the Simulation Interaction Diagram tool available in Desmond (The University of Queensland Research Computing Centre). 2024. Bunya supercomputer. Brisbane, Queensland, Australia. https://dx.doi.org/10.48610/wf6c-qy55).47–493 Result and discussion
3.1 Chemistry
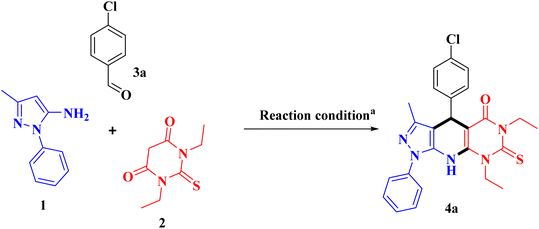
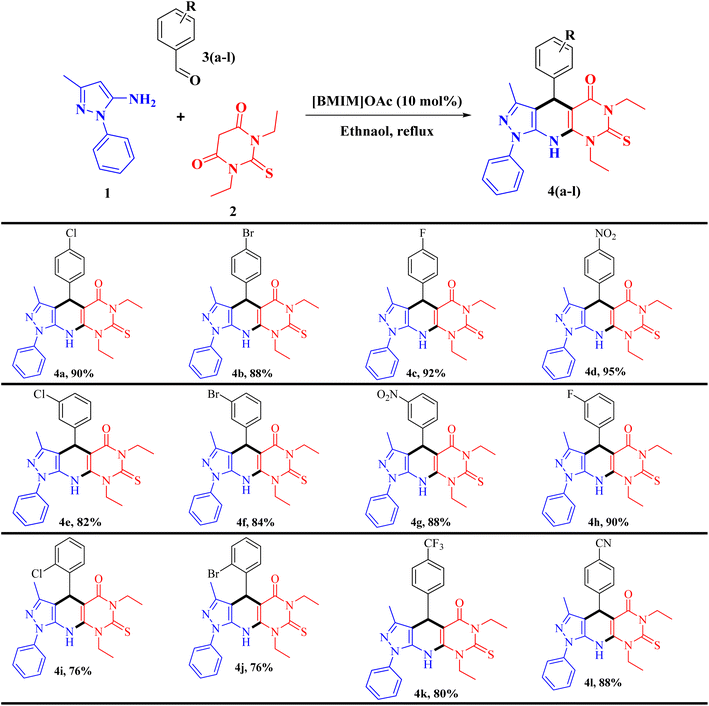
At the beginning of our experiment, we chose 5-amino-3-methyl-1-phenyl pyrazole (1, 1.0 mmol), 1,3-diethylthiobarbituric acid (2, 1 mmol), and p-chlorobenzaldehyde (3, 1 mmol) as model substrates to react in one-pot process. To determine the optimal reaction conditions for reaction, we tested different solvents and catalysts. Our first attempt involved heating the reactants in ethanol without using any catalyst for 24 hours, but it did not result in the desired product (entry 1, Table 1). The model reaction was tested with several different catalysts, as shown in Table 1. Among these, [BMIM]OAc at (10 mol%) in ethanol worked best, leading to full conversion of the starting materials, utilizing these best conditions, we did multicomponent reactions with compound 1, 2, and 3(a–l) to check how well and wide this method works. Aromatic aldehydes with electron-withdrawing groups reacted smoothly, giving isolated products with moderate to high yields between 76% to 95%. Electron-donating aromatic aldehydes (e.g., –Me, –OMe, –OH) were also screened; however, no desired product formation was observed, likely due to reduced electrophilicity of the carbonyl group. Therefore, the study was restricted to electron-withdrawing aromatic aldehydes, which afforded high yields. The range of substrate used is detailed in Table 2.| Entry | Catalyst (mol%) | Solvent | Temperature (°C) | Time (min) | Yieldb (%) |
|---|---|---|---|---|---|
| a Reaction conditions: 5-amino-3-methyl-1-phenyl pyrazole 1 (1.0 mmol), 1,3-diethylthiobarbituric acid 2 (1.0 mmol), and p-chlorobenzaldehyde 3a (1.0 mmol).b Isolated yield. | |||||
| 1 | — | Ethanol | Reflux | — | — |
| 2 | p-TSA (20) | Ethanol | Reflux | 180 | 77 |
| 3 | p-TSA (20) | Acetonitrile | Reflux | 160 | 82 |
| 4 | p-TSA (20) | N,N′-Dimethyl formamide | Reflux | 160 | 85 |
| 5 | L-Proline (20) | Ethanol | Reflux | 200 | 68 |
| 6 | L-Proline (20) | Acetonitrile | Reflux | 210 | 59 |
| 7 | — | Acetic acid | Reflux | 140 | 80 |
| 8 | [BMIM]Cl (20) | Ethanol | Reflux | 120 | 87 |
| 9 | [BMIM]Cl (10) | Ethanol | Reflux | 150 | 75 |
| 10 | [BMIM]OAc (20) | Ethanol | Reflux | 90 | 91 |
| 11 | [BMIM]OAc (15) | Ethanol | Reflux | 90 | 90 |
| 12 | [BMIM]OAc (10) | Ethanol | Reflux | 95 | 90 |
| a Reaction conditions: 5-amino-3-methyl-1-phenyl pyrazole 1 (1.0 mmol), 1,3-diethylthiobarbituric acid 2 (1.0 mmol), and aromatic aldehyde 3(a–l) (1.0 mmol); [BMIM]OAc (10 mol%), 4 mL ethanol, reflux. |
|---|
 |
As an example, the synthesis of compound 4a was confirmed by seeing a signal singlet at 5.37 ppm in the 1H-NMR spectrum. The key points were that there were no signals for the methylene protons from the 1,3-diethylthiobarbituric acid and no signal for aldehyde proton. Also, the lack of a signal for the aldehyde carbon at 200 ppm in the 13C-NMR spectrum helped to prove that the compound was formed.
In our recycling experiments, a model reaction was used to assess the catalytic performance of [BMIM]OAc. After confirming reaction completion via TLC, 5 mL of distilled water was added to precipitate the product, which was then collected by filtration. To recover the catalyst and eliminate water, the filtrate was concentrated under reduced pressure using rotary evaporator at 70–80 °C. this process was carried out for three additional cycles using the recover catalyst. A comparison between the third and the initial run indicated a decrease in product yield, as shown in Fig. 2.
3.2 Biological outcomes
| Compound | S. aureus | B. subtilis | E. coli | C. albicans | A. niger |
|---|---|---|---|---|---|
| 4a | 32 ± 1.5 | 64 ± 2.0 | 8 ± 2.1 | 64 ± 3.0 | 128 ± 3.4 |
| 4b | 16 ± 1.0 | 32 ± 1.3 | 8 ± 2.2 | 32 ± 2.0 | 64 ± 2.5 |
| 4c | 8 ± 0.9 | 16 ± 1.1 | 32 ± 1.5 | 32 ± 1.8 | 64 ± 2.0 |
| 4d | 16 ± 1.2 | 32 ± 1.4 | 16 ± 1.8 | 32 ± 1.9 | 64 ± 2.3 |
| 4e | 8 ± 0.8 | 16 ± 1.0 | 32 ± 1.2 | 32 ± 1.5 | 64 ± 2.1 |
| 4f | 16 ± 1.3 | 32 ± 1.5 | 8 ± 2.0 | 32 ± 2.1 | 64 ± 2.6 |
| 4g | 32 ± 1.4 | 32 ± 1.6 | 64 ± 2.3 | 64 ± 2.5 | 128 ± 3.0 |
| 4h | 16 ± 1.2 | 32 ± 1.4 | 32 ± 1.9 | 32 ± 2.0 | 64 ± 2.3 |
| 4i | 64 ± 2.1 | 64 ± 2.4 | 128 ± 3.2 | 32 ± 3.0 | 128 ± 3.5 |
| 4j | 16 ± 1.0 | 32 ± 1.2 | 64 ± 2.0 | 32 ± 2.1 | 64 ± 2.5 |
| 4k | 32 ± 1.3 | 32 ± 1.5 | 64 ± 2.1 | 16 ± 2.4 | 128 ± 3.2 |
| 4l | 16 ± 1.1 | 32 ± 1.3 | 32 ± 1.8 | 8 ± 2.0 | 64 ± 2.2 |
| Tetracycline | 36 ± 0.02 | 72 ± 0.05 | 16 ± 0.6 | — | — |
| Chloramphenicol | 38.8 ± 0.04 | 88.3 ± 0.05 | 42.5 ± 1.6 | ||
| Fluconazole | 64.0 ± 0.05 | 98.6 ± 0.06 | |||
| Miconazole | — | — | — | 126 ± 0.2 | 84.5 ± 0.8 |
| Compound | IC50 (µM) |
|---|---|
| 4a | 102 ± 3.5 |
| 4b | 60 ± 2.8 |
| 4c | 45 ± 2.1 |
| 4d | 55 ± 2.5 |
| 4e | 42 ± 1.9 |
| 4f | 58 ± 2.7 |
| 4g | 80 ± 3.0 |
| 4h | 65 ± 2.6 |
| 4i | 100 ± 3.7 |
| 4j | 50 ± 2.3 |
| 4k | 70 ± 2.9 |
| 4l | 55 ± 2.4 |
| Doxorubicin | 5 ± 0.4 |
The anticancer activity of compounds 4(a–l) was evaluated using the MTT assay, and the IC50 values are summarized in Table 4. Among the tested derivatives, 4e (42 ± 1.9 µM) and 4c (45 ± 2.1 µM) exhibited the highest cytotoxic activity, followed closely by 4j (50 ± 2.3 µM). Other compounds, including 4b, 4d, 4f, and 4h, displayed moderate activity with IC50 values ranging from 55–65 µM, whereas 4a and 4i were comparatively less potent (IC50 > 100 µM). In comparison, the standard drug doxorubicin showed a markedly higher potency with an IC50 of 5 ± 0.4 µM, highlighting its superior cytotoxic effect. Importantly, cytotoxic evaluation against HEK 293 normal human kidney cells revealed that most compounds exhibited low toxicity (IC50 > 50 µM), suggesting a favorable safety margin. Notably, halogenated analogues such as 4c and 4e demonstrated moderate cytotoxicity (∼40–50 µM) but remained significantly less toxic than doxorubicin, underscoring their potential as selective anticancer leads.
3.3 Molecular docking
In case of the molecular docking against the bacterial target, the results showed that the compounds 4c, 4e, and 4f possessed moderate binding affinities towards Tet repressor class D (PDB ID: 2VKE) with Glidescores of −4.378, −4.672, and −4.704 kcal mol−1 (Fig. 3(A)–(C), respectively), compared to tetracycline (co-crystallized ligand) at −5.108 kcal mol−1 (Fig. 3(D)). All three of 4c, 4e, and 4f showed a common H-bond interaction with the Hie100 (deprotonated form of His) residue (Table 5). Also, these compounds and tetracycline showed mutual polar contacts with Hie100, Gln109, Gln116, Ser138, and Hie139. Additionally, these three compounds showed common hydrophobic interactions with Leu60, Phe86, Pro105, Val113, Ile134, and Leu142 residues. All three compounds, as well as tetracycline, superimposed very well inside the binding pocket of Tet repressor class D.| Compound | Docking score (kcal mol−1) | H-bond | Hydrophobic interaction | Polar bond |
|---|---|---|---|---|
| PDB ID: 2VKE | ||||
| 4c | −4.378 | Hie100 | Leu60, Phe86, Leu101, Pro105, Val113, Leu117, Leu131, Ile134, and Leu142 | Hie100, Thr103, Gln109, Gln116, Ser135, Ser138, and Hie139 |
| 4e | −4.672 | Hie100 | Leu60, Phe86, Leu101, Pro105, Val113, Leu117, Leu131, Ile134, and Leu142 | Hie64, Hie100, Thr103, Gln109, Gln116, Ser135, Ser138, and Hie139 |
| 4f | −4.704 | Hie100 | Leu60, Phe86, Leu101, Pro105, Val113, Leu117, Leu131, Ile134, and Leu142 | Hie64, Ser67, Hie100, Thr103, Gln109, Gln116, Ser138, and Hie139 |
| Tetracycline | −5.108 | — | Leu60, Phe86, Pro105, Val113, Val137, Ile134, and Leu142 | Hie64, Ser67, Asn82, Hie100, Gln109, Thr112, Gln116, Ser135, Ser138, and Hie139 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| PDB ID: 5TZ1 | ||||
| 4c | −7.627 | — | Phe105, Tyr118, Leu121, Phe126, Ile131, Tyr132, Leu139, Phe228, Leu300, Ile304, Leu376, Ile379, Phe463, Cys470, Ile471, Met508, and Val509 | Thr122, Thr311, and His468 |
| 4e | −7.985 | Tyr132 | Leu121, Ile131, Tyr132, Leu139, Phe228, Ile304, Pro375, Leu376, Phe463, Cys470, Ile471, Phe475, Ala476, and Met508 | Thr311 and His468 |
| 4f | −6.969 | — | Tyr118, Leu121, Phe126, Ile131, Tyr132, Phe228, Leu300, Ile304, Leu370, Met374, Pro375, Leu376, Ile379, Pro462, Phe462, Phe463, Cys470, Ile471, Ala476, and Met508 | Thr311 and His468 |
| Fluconazole | −7.589 | Tyr132 | Tyr118, Leu121, Phe126, Ile131, Tyr132, Phe228, Phe233, Leu376, Ile379, Phe380, Met508, and Val509 | Thr122, Thr311, and Thr378 |
| Miconazole | −9.792 | Tyr132, Cys470 | Tyr118, Leu121, Phe126, Ile131, Tyr132, Leu139, Phe228, Phe233, Leu300, Ile304, Ile379, Cys470, Ile471, and Met508 | Thr122, Thr122, and Thr311 |
In case of the molecular docking against the fungal target, the results showed that the compounds 4c, 4e, and 4f possessed moderate binding affinities towards sterol 14-alpha demethylase (PDB ID: 5TZ1) with Glidescores of −7.627, −7.985, and −6.969 kcal mol−1 (Fig. 4(A)–(C), respectively), compared to the standard drugs, i.e., fluconazole and miconazole, at −7.589 and −9.792 kcal mol−1 (Fig. 4(D) and (E), respectively). Among the three compounds, 4e showed a mutual H-bond interaction with Tyr132 with both the standard drugs (Table 5). Also, all three compounds showed a common polar contact with Thr311 and His468, while all of them possessed a single mutual polar contact with Thr311 residue, with the standard drugs. All 5 compounds (4c, 4e, 4f, and the standard drugs) showed common hydrophobic interactions with Leu121, Ile131, Tyr132, Phe228, and Met508. All three compounds, as well as the standard drugs, superimposed moderately inside the binding pocket of the target.
3.4 Molecular dynamics (MD) simulations
MD studies of the 4f-Tet repressor class D (PDB ID: 2VKE) complex and 4f-sterol 14-alpha demethylase (PDB ID: 5TZ1) were done on the top Glide Score-containing complex. For the 4f-Tet repressor class D complex (PDB ID: 2VKE), the Root Mean Square Deviation (RMSD) remained below 2 Å throughout the 200 ns molecular dynamics simulation (Fig. 5(A)), indicating moderate structural stability within the tetracycline-binding site of the Tet repressor class D. Analysis of the protein–ligand interaction diagram revealed that His100, Pro105, Tyr132, and Leu142 contribute significantly to complex stability (Fig. 5(B) and (C)). Among these, Pro105, Tyr132, and Leu142 predominantly participate in hydrophobic interactions at the active site, each with an interaction fraction exceeding 0.25. Furthermore, His100 primarily forms hydrogen bonds and water bridges, both persisting for more than 15% of the simulation time. Overall, the in silico results suggest that His100, Pro105, and Leu142 are key residues influencing antibacterial activity, consistent with their involvement in the molecular docking interactions of the standard tetracycline compound.For the 4e-sterol 14-alpha demethylase complex (PDB ID: 5TZ1), the Root Mean Square Deviation (RMSD) remained below 1 Å during the 200 ns molecular dynamics simulation (Fig. 6(A)), indicating a highly stable complex within the tetrazole-based antifungal binding site of sterol 14-alpha demethylase. According to the protein–ligand interaction analysis, Tyr132, Phe463, His468, and Ala476 play key roles in maintaining the stability of the complex (Fig. 6(B) and (C)). Among these, Phe463 and Ala476 primarily form hydrophobic interactions at the active site, each with an interaction fraction exceeding 0.5. Additionally, Tyr132 and His468 contribute major water–bridge interactions, persisting for more than 60% of the simulation time. There is no significant H-bond interaction (only Thr311; interaction fraction of <0.1) reported here. The conformational stability of the docked complexes was further assessed using MD simulations. Representative snapshots at different simulation time points are shown in Fig. 7.
 | ||
| Fig. 7 Results of MD simulations of 4f-2VKE and 4e-5TZ1 at 1st (A), 2500th (B), 5000th (C), 7500th (D), 10002nd (E) frames of MD simulations. | ||
According to the in silico findings, Tyr132 and Leu376 appear to play crucial roles in antifungal activity, as these interactions were consistently observed in the molecular docking results for the reference compounds Fluconazole and Miconazole.
4 Conclusion
In summary, we have established a green, efficient, and recyclable ionic-liquid-mediated multicomponent strategy for the synthesis of pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidin-5-one derivatives using [BMIM]OAc as a catalyst. This protocol provides several advantages over previously reported methods, including mild reaction conditions, shorter reaction times, excellent product yields, and simple catalyst recovery and reusability, thereby aligning closely with green chemistry principles. The antimicrobial screening studies revealed that compounds 4a, 4b, 4c, 4e, 4f, and 4l exhibited potent activity, outperforming standard reference drugs in selected microorganisms, while the majority of synthesized molecules demonstrated low cytotoxicity, indicating a favourable therapeutic window. Furthermore, molecular docking and MD simulations confirmed strong and stable binding interactions, supporting their biological potential. Overall, this research provides a valuable foundation for future applications of ionic-liquid-assisted multicomponent reactions in medicinal chemistry and supports the further exploration of these heterocycles for potential antimicrobial and anticancer therapeutic development.Author contributions
Savan S. Bhalodiya: method development, investigation, validation, spectral analysis, writing an original draft, Mehul P. Parmar: writing – review editing, spectral analysis, Shana Balachandran: investigation, biological study, writing – review & editing, formal analysis, Chirag D. Patel: writing – review & editing, formal analysis, Arijit Nandi: computational study, writing – review & editing, visualization, formal analysis, Anwesha Das: computational study, writing – review & editing, formal analysis, Madan Kumar Arumugam: biological study, writing – review & editing, formal analysis, Hitendra M. Patel: writing – review & editing, supervision, method development, investigation, formal analysis, conceptualization.Conflicts of interest
The authors declare no conflict of interest.Data availability
The data supporting this article has been included as part of the supporting information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra09581f.Acknowledgements
We would like to extend our heartfelt gratitude to the Department of Chemistry, Sardar Patel University, Vallabh Vidyanagar for providing us with the necessary research facilities. SSB is thankful to the CSIR-NET fellowship (Reference No. Nov/06/2020(i)EU-V). MPP is thankful to the Knowledge Consortium of Gujarat for SHODH fellowship (Student Reference No. 2021016434). CDP is thankful to National fellowship for scheduled Tribe (Award No. 202223-NFST-GUJ-00003). This work was supported by resources provided by The University of Queensland Research Computing Centre's Bunya supercomputer, with funding from The University of Queensland, Brisbane, Australia.References
- E. Kabir and M. Uzzaman, Results Chem., 2022, 4, 100606 Search PubMed.
- H. D. Nguyen and M.-S. Kim, Comput. Biol. Chem., 2023, 104, 107872 CrossRef CAS.
- S. Chigurupati, A. Al-murikhy, S. A. Almahmoud, Y. Almoshari, A. Saber Ahmed, S. Vijayabalan, S. Ghazi Felemban and V. Raj Palanimuthu, Saudi J. Biol. Sci., 2022, 29, 854–859 CrossRef CAS.
- P. J. Patel, S. G. Patel, D. B. Upadhyay, L. Ravi, A. Dhanasekaran and H. M. Patel, RSC Adv., 2023, 13, 24466–24473 RSC.
- A. P. Taylor, R. P. Robinson, Y. M. Fobian, D. C. Blakemore, L. H. Jones and O. Fadeyi, Org. Biomol. Chem., 2016, 14, 6611–6637 RSC.
- K. R. A. Abdellatif and R. B. Bakr, Med. Chem. Res., 2021, 30, 31–49 Search PubMed.
- S. Schenone, M. Radi, F. Musumeci, C. Brullo and M. Botta, Chem. Rev., 2014, 114, 7189–7238 CrossRef CAS.
- K. Kandhasamy, R. R. Surajambika and P. K. Velayudham, Med. Chem., 2024, 20(3), 293–310 CrossRef CAS.
- S. S. Bhalodiya, M. P. Parmar, D. B. Upadhyay, C. D. Patel, D. P. Vala, D. Rajani and H. M. Patel, Results Chem., 2024, 7, 101304 CrossRef CAS.
- M. P. Parmar, D. P. Vala, S. S. Bhalodiya, D. B. Upadhyay, C. D. Patel, S. G. Patel, S. R. Gandholi, A. H. Shaik, A. D. Miller, J. Nogales, S. Banerjee, J. M. Padrón, N. Amri, N. K. Kandukuri and H. M. Patel, RSC Adv., 2024, 14, 9300–9313 RSC.
- S. G. Patel, D. B. Upadhyay, N. V. Shah, M. P. Parmar, P. J. Patel, A. Malik, R. K. Sharma and H. M. Patel, RSC Sustain., 2024, 2, 1128–1141 RSC.
- P. J. Patel, R. M. Vala, S. G. Patel, D. B. Upadhyay, D. P. Rajani, F. Damiri, M. Berrada and H. M. Patel, J. Mol. Struct., 2023, 1285, 135467 Search PubMed.
- R. Javahershenas, J. Han, M. Kazemi and P. J. Jervis, ChemistrySelect, 2024, 9, e202401496 Search PubMed.
- M. P. Parmar, S. Kashyap, D. P. Vala, S. S. Bhalodiya, C. D. Patel, J. Nogales, A. Nandi, S. Banerjee and H. M. Patel, Eur. J. Med. Chem., 2025, 297, 117914 CrossRef CAS PubMed.
- D. M. Patel, P. J. Patel and H. M. Patel, Eur. J. Org Chem., 2022, 2022, e202201119 Search PubMed.
- R. Javahershenas, J. Han, M. Kazemi and P. J. Jervis, ChemistryOpen, 2024, 13, e202400185 CrossRef CAS PubMed.
- R. Javahershenas and S. Nikzat, RSC Adv., 2023, 13, 16619–16629 Search PubMed.
- S. S. Bhalodiya, M. P. Parmar, C. D. Patel, S. G. Patel, D. P. Vala, N. Suresh, B. Jayachandran, M. Kumar Arumugam, M. Narayan and H. M. Patel, ChemMedChem, 2025, 20, e202400595 CrossRef CAS.
- M. P. Parmar, A. Das, D. P. Vala, S. S. Bhalodiya, C. D. Patel, S. Balachandran, N. K. Kandukuri, S. Kashyap, A. N. Khan, A. González-Bakker, M. K. Arumugam, J. M. Padrón, A. Nandi, S. Banerjee and H. M. Patel, ACS Omega, 2025, 10, 7013–7026 CrossRef CAS.
- D. B. Upadhyay, J. A. Mokariya, P. J. Patel, S. G. Patel, A. Das, A. Nandi, J. Nogales, N. More, A. Kumar, D. P. Rajani, M. Narayan, J. Kumar, S. Banerjee, S. K. Sahoo and H. M. Patel, Arch. Pharmazie, 2024, 357, 2300673 CrossRef CAS.
- M. A. P. Martins, C. P. Frizzo, D. N. Moreira, N. Zanatta and H. G. Bonacorso, Chem. Rev., 2008, 108, 2015–2050 CrossRef CAS.
- N. Isambert, M. d. M. S. Duque, J.-C. Plaquevent, Y. Génisson, J. Rodriguez and T. Constantieux, Chem. Soc. Rev., 2011, 40, 1347–1357 RSC.
- S. Rajamanickam and B. Dam, in Handbook of Ionic Liquids, 2024, pp. 419–442, DOI:10.1002/9783527839520.ch21.
- M. Zakeri, M. M. Nasef and E. Abouzari-Lotf, J. Mol. Liq., 2014, 199, 267–274 CrossRef CAS.
- J. B. Bharate, S. B. Bharate and R. A. Vishwakarma, ACS Comb. Sci., 2014, 16, 624–630 CrossRef CAS PubMed.
- F. Shirini, M. S. N. Langarudi, N. Daneshvar, M. Mashhadinezhad and N. Nabinia, J. Mol. Liq., 2017, 243, 302–312 CrossRef CAS.
- O. G. Jolodar, F. Shirini and M. Seddighi, Chin. J. Catal., 2017, 38, 1245–1251 CrossRef CAS.
- A. Arias-Gómez, A. Godoy and J. Portilla, Molecules, 2021, 26(9), 2708 CrossRef.
- P. Yadav and K. Shah, Chemical Biology & Drug Design, 2021, vol. 97, pp. 633–648 Search PubMed.
- D. J. Baillache and A. Unciti-Broceta, RSC Med. Chem., 2020, 11, 1112–1135 RSC.
- A. Ballesteros-Casallas, M. Paulino, P. Vidossich, C. Melo, E. Jiménez, J.-C. Castillo, J. Portilla and G. P. Miscione, Eur. J. Med. Chem. Rep., 2022, 4, 100028 CAS.
- H. C. Patel, M. S. Patel, J. N. Parekh, D. D. Chudasama, P. Dalwadi, A. Kunjadiya, V. Bhatt, K. M. Modi, C. N. Patel and K. R. Ram, J. Biomol. Struct. Dyn., 2025, 43, 3467–3490 CrossRef CAS.
- A. S. Hassan, N. M. Morsy, H. M. Awad and A. Ragab, J. Iran. Chem. Soc., 2022, 19, 521–545 CrossRef CAS.
- E. H. El-Sayed and K. S. Mohamed, Polycyclic Aromat. Compd., 2021, 41, 1077–1093 CrossRef CAS.
- S. Deshmukh, K. Dingore, V. Gaikwad and M. Jachak, J. Chem. Sci., 2016, 128, 1459–1468 CrossRef CAS.
- M. Faisal, A. Saeed, S. Hussain, P. Dar and F. A. Larik, J. Chem. Sci., 2019, 131, 70 CrossRef.
- N. Abbas, P. M. G. Swamy, P. Dhiwar, S. Patel and D. Giles, Pharm. Chem. J., 2021, 54, 1215–1226 CrossRef CAS.
- G. Rajitha, V. Ravibabu, G. Ramesh and B. Rajitha, Res. Chem. Intermed., 2016, 42, 1989–1998 CrossRef CAS.
- P. Rathee, R. Tonk, A. Dalal, M. Ruhil and A. Kumar, Cell. Mol. Biol., 2016, 62, 4172 Search PubMed.
- A. K. Oraby, R. Khaled, M. Abdelgawad, K. Attia and P. Georghiou, Int. J. Pharm. Chem., 2016, 6, 100–106 Search PubMed.
- A. Bazgir, M. M. Khanaposhtani and A. A. Soorki, Bioorg. Med. Chem. Lett., 2008, 18, 5800–5803 CrossRef CAS PubMed.
- S. P. Satasia, P. N. Kalaria and D. K. Raval, Org. Biomol. Chem., 2014, 12, 1751–1758 RSC.
- M. Daraie, M. M. Heravi, M. Mirzaei and N. Lotfian, Appl. Organomet. Chem., 2019, 33, e5058 CrossRef.
- M. A. Ghasemzadeh, B. Mirhosseini-Eshkevari and S. Sanaei-Rad, Polyhedron, 2022, 212, 115588 Search PubMed.
- M. M. Heravi and M. Daraie, Molecules, 2016, 21(4), 441 CrossRef PubMed.
- M. Daraie and M. M. Heravi, Arkivoc, 2016, 2016, 328–338 Search PubMed.
- Y. K. Mahdi, M. R, D. Kumar, A. Nandi, A. Das, Y. A. Atia, R. Ranjan and Y. N. Dey, Curr. Trends Biotechnol. Pharm., 2023, 17, 1496–1505 Search PubMed.
- K. J. Bowers, E. Chow, H. Xu, R. O. Dror, M. P. Eastwood, B. A. Gregersen, J. L. Klepeis, I. Kolossvary, M. A. Moraes, F. D. Sacerdoti, J. K. Salmon, Y. Shan and D. E. Shaw, Presented in Part at the Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, Florida, 2006 Search PubMed.
- A. Nandi, N. Mandal, A. Das and Y. N. Dey, J. Biol. Regul. Homeostatic Agents, 2024, 38, 2055–2067 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |