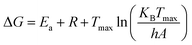Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Novel amide-based macrocyclic Co(II) complexes: correlating structural, computational, and biological properties through DFT and docking
Subhashac,
Manish Kumarb,
Vandanac,
Annu Devic and
Ashu Chaudhary *c
*c
aDepartment of Chemistry, Smt. Devkiba Mohansinhji Chauhan College of Commerce and Science, UT of DNH & DD, Silvassa 396230, India
bDepartment of Chemistry, Maharshi Dayanand University, Rohtak 124001, Haryana, India
cDepartment of Chemistry, Kurukshetra University, Kurukshetra 136119, Haryana, India. E-mail: ashuchaudhary21@gmail.com; ashuchaudhary@kuk.ac.in; Tel: +91-9729864551
First published on 14th January 2026
Abstract
A series of Co(II) complexes incorporating functionalized macrocyclic scaffolds, designated as [Co(N4O4MacL1)Cl2]–[Co(N4O4MacL3)Cl2], were synthesized using three structurally related macrocyclic ligands (N4O4MacL1–N4O4MacL3), each featuring an amide-based tetradentate donor framework. The complexes were comprehensively characterized by elemental analysis, FTIR spectroscopy, magnetic susceptibility measurements, UV-visible spectroscopy, 1H and 13C NMR spectroscopy, and powder X-ray diffraction. Spectroscopic and analytical findings confirm that the ligands act as monoanionic tetradentate donors, coordinating through nitrogen and oxygen donor sites. The magnetic and electronic spectral data support an octahedral coordination geometry around the Co(II) center in all complexes. Thermal stability and kinetic parameters were assessed through thermogravimetric analysis, employing the Coats-Redfern and Flynn-Wall-Ozawa (FWO) models to determine activation energies and thermodynamic parameters. Moreover, density functional theory (DFT) calculations provided insights into optimized geometries, bond parameters, and frontier molecular orbital characteristics, which corroborate the experimental observations. The biological activities of the ligands and their Co(II) complexes were evaluated through in vitro antimicrobial assays against fungal strains A. niger and C. albicans, and bacterial strains B. subtilis, S. aureus, E. coli, and S. typhi. Additionally, antioxidant activity assessed by the DPPH free-radical scavenging assay revealed that [Co(N4O4MacL3)Cl2] demonstrated the highest radical-quenching efficiency. Among the complexes studied, [Co(N4O4MacL2)Cl2] displayed the most promising antimicrobial activity with minimal cytotoxic effects. Molecular docking studies further validated the biological results, indicating favourable interactions of the complexes with relevant biomolecular targets. Overall, these findings highlight the therapeutic potential of amide-containing macrocyclic ligands and their cobalt(II) complexes as promising candidates for antimicrobial applications, selective cytotoxic behaviour, and effective antioxidant activity.
1 Introduction
Bioinorganic chemistry has emerged as an important and expanding branch of inorganic chemistry, tracing its origins to the development of cisplatin one of the first metal-based anticancer drugs. Cancer, characterized by the uncontrolled proliferation of abnormal cells, leads to life-threatening diseases. As reported by the World Health Organization, it stands as the second most common cause of illness and death worldwide, following cardiovascular diseases.1 Although significant advancements have been made in cancer detection, treatment, and prevention, it continues to be a leading cause of mortality across the globe. Most cancer drugs currently in use are not specifically targeted toward tumour cells; instead, they exhibit general cytotoxic effects that also harm healthy cells. Consequently, there is an increasing need to design new and more selective therapeutic agents for cancer treatment.2–4 The search for novel compounds with antitumor potential remains a crucial focus of pharmaceutical research. Metal complexes containing oxygen- and nitrogen-donor ligands have shown promising biological activities, including antibacterial, antifungal, and antitumor effects, along with various biochemical, pharmacological, and clinical properties.5–10 These biologically active substances include metal ions, coordination compounds, complex ions, and other inorganic molecules.11,12 The strong chelating ability of macrocyclic ligands plays an important role in their antioxidant behaviour, which can contribute to the development of transition-metal-based anticancer agents.13–16 Moreover, density functional theory (DFT) has become an increasingly valuable tool in theoretical studies of metal complexes, providing insights into their stereochemical structures and electronic properties.17–20 The remarkable biological responsiveness of macrocyclic compounds and their ability to chelate transition metal ions present in biological systems have attracted significant scientific attention. These compounds exhibit strong chelating tendencies, allowing them to form a wide range of coordination geometries from bidentate to multidentate through donor systems such as ONO, and NO. Metal complexes of transition metals including Mn(II), Cu(II), Cr(III), and Fe(III) have demonstrated notable roles in biological processes, particularly exhibiting antioxidant, antimicrobial, DNA-binding, and cytotoxic properties.21–24 Consequently, macrocyclic ligands containing the donor framework (N4O4MacL1–N4O4MacL2) and their corresponding metal complexes are anticipated to possess significant and diverse biological activities.2 Experimental section
2.1 Material and methods
All culture media utilized in this study were procured from HiMedia (India) and used as supplied, without additional processing. High-purity analytical-grade solvents were employed directly, with no further purification steps. Elemental composition of the synthesized compounds was determined using an Elementar Vario EL III CHN analyzer at the Sophisticated Analytical Instrumentation Facility (SAIF). Fourier-transform infrared (FT-IR) spectra were recorded on a PerkinElmer Spectrum IR (Version 10.6.2) spectrophotometer using KBr pellets prepared in-house. Electron paramagnetic resonance (EPR) analyses were performed in both solid state and DMF medium at liquid nitrogen temperature on a Varian F-113 X-band spectrometer, employing TCE (g = 2.00277) as the ordinary SAIF, at, IIT Bombay, India. The cancer cell line employed in this investigation was kindly provided by a laboratory at the Indian Institute of Science (IISc), Bengaluru, India, and was maintained under standard culture conditions. Proton and carbon NMR spectra were obtained in DMSO-d6 using a JEOL 400 ECZS spectrometer. Electron-ionization mass spectra (EI-MS) for the synthesized ligands and cobalt(II) complexes were acquired on a XEVO G2-XS QTof high-resolution mass spectrometer to validate molecular weights and assess characteristic fragmentation behavior. UV-visible spectral measurements were carried out in DMSO using a PerkinElmer Lambda series UV-vis spectrophotometer. Powder X-ray diffraction (PXRD) data were collected on a PANalytical X'Pert Pro diffractometer. Thermal behavior and decomposition characteristics were evaluated via thermogravimetric analysis (TGA) using a Shimadzu Model 50 instrument.2.2 Design and execution of synthetic pathways
Following the same optimized protocol, cobalt complexes [Co(N4O4MacL2)Cl2] and [Co(N4O4MacL3)Cl2] were synthesized by substituting N4O4MacL1 with N4O4MacL2 (0.9615 g, 2 mmol) and N4O4MacL3 (1.0159 g, 2 mmol), respectively. Variation in ligand chain length facilitated controlled modulation of the macrocyclic cavity and metal coordination environment. The successful formation of the complexes was confirmed by characteristic spectroscopic features, including ligand-to-metal charge transfer bands and disappearance of free ligand functional group signals. To ensure high purity of the synthesized compounds, we adopted a combination of careful synthetic control and multiple purification steps. High purity levels were primarily achieved by optimizing reaction conditions, including reagent molar ratios, solvent selection, reaction time, and temperature, which minimized the formation of side products. After completion of the reaction, the crude product was subjected to appropriate purification techniques such as recrystallization, solvent–solvent extraction, and column chromatography depending on the compound's physicochemical properties. These steps ensured selective isolation of the target compound (Scheme 1).
2.3 DFT calculations
Quantum-chemical investigations of the tetra amide based macrocyclic ligands (N4O4MacL1–N4O4MacL3) and their cobalt(II) complexes were performed using the ORCA computational suite27 employing the density functional theory (DFT) framework. Geometry optimizations were conducted without imposing symmetry constraints.28 The hybrid B3LYP functional was used in combination with the def2-SVP basis set for ligand atoms, while the def2-TZVP basis set was applied to cobalt centers to ensure reliable treatment of transition-metal electronic environments. The B3LYP functional was selected because it is one of the most extensively benchmarked hybrid functionals for geometry optimization, electronic structure calculations, and prediction of molecular properties of coordination and organic compounds. Numerous studies demonstrate that B3LYP provides a reliable balance between computational cost and accuracy for systems comparable to those investigated in this work. The def2-SVP basis set was employed as it offers a well-validated split-valence description with polarization functions, enabling accurate geometry optimization while maintaining reasonable computational efficiency for molecules of moderate size. Test calculations using a larger def2-TZVP basis set on representative structures showed only marginal differences in optimized geometries and electronic energies, supporting the suitability of def2-SVP for full-scale calculations. Regarding dispersion and solvent corrections, the primary focus of our study was on gas-phase intrinsic electronic properties, for which B3LYP/def2-SVP provides dependable results. Nonetheless, dispersion-corrected (D3) and implicit solvent (CPCM) single-point energy calculations were additionally performed on the optimized geometries to verify the robustness of the computed parameters.After successful optimization, no imaginary frequencies were observed, confirming that the structures correspond to true energy minima. Frontier molecular orbital energies, including the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO), total electronic energies, and dipole moments were calculated to assess electronic characteristics and reactivity profiles. In addition, optimized geometrical parameters such as selected bond lengths, bond angles, and coordination metrics around the metal ion were examined to elucidate structural features and intra-ligand interactions. These theoretical insights complement the experimental findings, providing a deeper understanding of electronic distribution, stability, and molecular geometry in both the free ligands and their cobalt(II) complexes.
2.4 Biology
Nutrient agar medium was prepared using peptone (5 g), beef extract (5 g), sodium chloride (5 g), and agar–agar (20 g) in 1000 mL of distilled water. The sterilized medium was poured into Petri plates and allowed to solidify. Subsequently, ∼5.4 mL of sterile molten nutrient agar inoculated with the test culture (approximately 41 °C) was layered over the base medium to obtain a uniform lawn of microbial growth. Test samples were dissolved in petroleum ether to obtain concentrations of 30 and 50 ppm. Sterile Whatman no. 1 filter paper discs (5 mm diameter) were impregnated with the sample solutions, air-dried under aseptic conditions, and carefully placed on the inoculated agar plates. The plates were incubated at 29.4 °C for 22 h, after which the diameters of inhibition zones (in mm) were recorded to evaluate antibacterial activity. Ampicillin served as the positive control and was tested under identical conditions using the same solvent system. The antifungal efficacy of ligands (N4O4MacL1–N4O4MacL3) and their corresponding cobalt(II) complexes ([Co(N4O4MacL1)Cl2]–[Co(N4O4MacL3)Cl2]) was examined using the agar well diffusion method29 against pathogenic fungi Aspergillus niger and Candida albicans. Experimental conditions including medium composition, inoculum preparation, sample concentrations, and incubation parameters were maintained similar to the antibacterial protocol to ensure consistency. Zones of fungal growth inhibition were measured to determine antifungal potential.
2.5 Antioxidant activity
The antioxidant efficiency of the synthesized ligands and their cobalt(II) complexes was assessed using the DPPH (2,2-diphenyl-1-picrylhydrazyl) free-radical scavenging method. The assay was conducted spectrophotometrically using a UV-visible spectrophotometer. A 1 mM DPPH stock solution was freshly prepared in methanol,30 and serial dilutions of the test samples (0–60 mg mL−1) were prepared in DMSO. Ascorbic acid served as the reference antioxidant under identical experimental conditions.For each experiment, 1.5 mL of the test solution was combined with 3.5 mL of 0.1 mM DPPH solution in methanol. The reaction mixtures were vortexed briefly to ensure homogenization and incubated at room temperature in the dark for 30 minutes to allow complete interaction between the radical species and the test compounds. A control was prepared using DPPH solution without the sample. Following incubation, absorbance values were measured at 517 nm, corresponding to the characteristic absorbance maximum of DPPH. The decrease in absorbance relative to the control indicated free-radical scavenging capacity.31
The experimental protocol enabled quantitative determination and comparison of radical-scavenging performance among the synthesized ligands and their cobalt(II) derivatives.
2.6 Cytotoxicity
The in vitro cytotoxic potential of the synthesized Macrocyclic ligands (N4O4MacL1–N4O4MacL3) and their corresponding cobalt(II) complexes was evaluated against human breast carcinoma (MCF-7) and hepatocellular carcinoma (HepG-2) cell lines using the standard MTT colorimetric assay.32 MCF-7 and HepG-2 cells were routinely maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (P/S), and incubated at 37 °C in a humidified atmosphere containing 5% CO2. Cells were seeded into sterile 96-well plates at a density of approximately 1 × 104 cells per well and allowed to adhere overnight. Upon reaching ∼92–94% confluence, cells were treated with graded concentrations of the synthesized compounds (3.125–100.0 µM) along with colchicine, which served as a positive control. Untreated cells incubated under identical conditions were used as a negative control. Plates were then incubated for 46 h at 38 °C to allow compound cell interaction. All experiments were conducted in triplicate and results were expressed as mean ± standard error (SE). The cytotoxicity dose–response curves were generated using a four-parameter logistic (4 PL) non-linear regression model,33 which is widely accepted for sigmoidal dose–response analyses. The model is defined as:| Y = Bottom + (Top − Bottom)/[1 + (X/IC50)n] |
2.7 Molecular docking investigations
The Molecular docking has recently become a vital tool in the design and development of new drug candidates.34 Its primary purpose is to elucidate drug–nucleic acid or drug–protein interactions and to predict the preferred binding orientation of a ligand within a target biomolecule. In this study, AutoDock Vina,35 a rigid docking software, was employed to investigate the binding interactions of three Macrocyclic ligands (N4O4MacL1–N4O4MacL3). The crystal structures of all target receptors were obtained from the Protein Data Bank.The study aimed to predict the most favorable binding modes and the corresponding lowest binding energies of the ligands. Experimental results indicated that N4O4MacL2 exhibited notable in vitro antibacterial and antifungal activity, effectively inhibiting the growth of bacteria and fungi. Docking analyses revealed that all three ligands (N4O4MacL1–N4O4MacL3) showed significant interactions with proteins from E. coli, S. aureus, C. albicans, and B. subtilis. The binding energies (kcal mol−1) calculated at the active sites of these organisms provided a qualitative assessment of inhibitory potentimagal.
Notably, N4O4MacL2, which contains a para-substituted halogen on the benzene ring, exhibited stronger binding to E. coli proteins compared to streptomycin, suggesting that this substitution enhances the ligand's fit within the active site and contributes to its biological activity.36
3 Results and discussion
3.1 Common features
The physical, analytical, and spectroscopic characteristics of the ligands (N4O4MacL1–N4O4MacL3) and their divalent cobalt complexes are summarized in the experimental section (Table S1). The complexes are soluble in polar aprotic solvents such as DMF and DMSO and exhibit stability under ambient conditions. The experimentally determined elemental composition closely matches the calculated values, confirming the formation of the complexes in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 metal-to-ligand stoichiometry.37
1 metal-to-ligand stoichiometry.37
3.2 FT-IR characterization and metal–ligand binding features
The FT-IR spectra of the complexes provided the first indication of successful macrocyclic complex formation, as several characteristic peaks present in the spectra of the free ligands were absent in the complexes. The sharp band observed in the 3289–3302 cm−1 range in the ligand spectra was assigned to the cyclized, non-coordinated amide N–H stretch.38 Strong absorption bands between 1536–1582 cm−1, formed through an intramolecular condensation reaction between the –OH groups of the dicarboxylic acids and the –NH2 moieties of 1,8-diaminonaphthalene, leading to macrocyclic ring closure, further supported the formation of macrocyclic ligands. Additionally, four amide-associated bands appeared at 1663–1673, 1534–1552, 1245–1279, and 633–649 cm−1, consistent with those reported for previously characterized macrocyclic compounds (Fig. 1).39In the cobalt complexes, the amide band shifted slightly to 1679–1689 cm−1 compared to 1663–1673 cm−1 in the free ligands, indicating coordination via the nitrogen atoms of the macrocyclic moiety (Fig. S1–S3).41 A new band observed in the 429–455 cm−1 region was assigned to the ν(M–N) vibration, confirming metal–nitrogen coordination.40 Furthermore, bands appearing at 322–343 cm−1 were attributed to ν(M–Cl) vibrations (Table 1). The combined spectral signatures demonstrate that cobalt(II) is hexacoordinated through four amide nitrogen donors and two coordinating chloride anions.
| Complexes | ν(–NH) | Amide | ν(Co–N) | ν(Co–Cl) | |||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | ||||
| [N4O4MacL1] | 3292 | 1673 | 1534 | 1245 | 633 | — | — |
| [N4O4MacL2] | 3284 | 1668 | 1546 | 1269 | 638 | — | — |
| [N4O4MacL3] | 3275 | 1663 | 1552 | 1279 | 649 | — | — |
| [Co(N4O4MacL1)Cl2] | 3270 | 1689 | 1565 | 1277 | 648 | 429 | 322 |
| [Co(N4O4MacL2)Cl2] | 3259 | 1687 | 1581 | 1299 | 660 | 447 | 331 |
| [Co(N4O4MacL3)Cl2] | 3254 | 1679 | 1580 | 1309 | 661 | 455 | 343 |
3.3 1H and 13C-NMR spectra
Proton NMR spectra provided additional confirmation for the successful synthesis of the amide-containing Macrocyclic ligands (N4O4MacL1–N4O4MacL3).41 The spectra, recorded in DMSO-d6 at room temperature (Fig. 3), indicate the stability of the ligand framework. The absence of signals corresponding to the primary amino protons of 1,8-diaminonaphthalene in the 1H NMR spectra suggests that the expected Macrocyclic structures were successfully formed. The presence of a broad peak between δ 9.76–10.11 ppm, assignable to amide –NH resonances, confirms the formation of amide linkages through condensation of amine groups with carboxyl moieties.42The singlet signals at δ 3.75, 3.12, and 3.39 ppm in N4O4MacL1, N4O4MacL2, and N4O4MacL3, respectively, are attributed to the methylene protons (–CH2–) of the condensed diacid units (adipic, succinic, and glutaric acids). Additionally, slightly broad signals for amine protons appear between δ 6.13–6.52 ppm, while aromatic protons resonate in the δ 7.23–7.79 ppm range (Fig. 2).43 Characteristic aromatic proton resonances are also present, as detailed in Table 2. Further validation of Macrocycle formation and amide linkage incorporation is furnished by the 13C NMR spectral data. Broad peaks were observed at δ 165.36–168.85 ppm for amide carbons, δ 116.05–132.54 ppm for aromatic carbons, and δ 45.57–49.72 ppm for aliphatic carbons (Table 3 and Fig. 3),44 confirming the expected chemical environment of the macrocyclic frameworks.
| Ligands | (CO–NH) (m) | 1,4-C6H4 (m) | 1,2-C6H4 (m) | –CH2 (m) |
|---|---|---|---|---|
| [N4O4MacL1] | 9.76 | 7.35–7.75 | 6.30–6.41 | 3.75–3.95 |
| [N4O4MacL2] | 9.89 | 7.23–7.79 | 6.22–6.52 | 3.12–3.35 |
| [N4O4MacL3] | 10.11 | 7.38–7.57 | 6.13–6.38 | 3.39–3.49 |
| Compound | (CO–NH) | 1,4-C6H4 | 1,2-C6H4 | –CH2 (m) |
|---|---|---|---|---|
| [N4O4MacL1] | 167.13 | 116.05, 116.74 | 127.36, 132.26 | 45.57 |
| [N4O4MacL2] | 165.36 | 116.25, 117.08 | 127.32, 132.54 | 47.53 |
| [N4O4MacL3] | 168.85 | 116.87, 119.58 | 130.41, 132.15 | 49.72 |
3.4 Mass spectral
The ligands and their Co(II) complexes were characterized by electrospray ionization mass spectrometry (ESI-MS) in DMSO solution. Confirmation of the ligand's molecular formula was obtained from its positive-ion ESI mass spectrum, which displayed a prominent molecular ion peak at m/z 480, corresponding to the molecular weight of C28H24 (calculated m/z 380.3413). The spectra of the ligands (N4O4MacL1–N4O4MacL3) and the divalent cobalt complexes exhibited prominent molecular ion peaks represented as M+ or (M + H)+.In addition to the molecular ion peaks, the spectra displayed fragments resulting from the loss of phenyl or benzyl substituents. Several other characteristic peaks were also observed in the mass spectra, confirming the composition of the compounds. For example, the fragmentation pattern of N4O4MacL2 showed a molecular ion peak at m/z 480 (C26H20). The ligands N4O4MacL1, N4O4MacL3 and the corresponding Co(II) complexes ([Co(N4O4MacL1)Cl2]–[Co(N4O4MacL3)Cl2]) exhibited molecular ion peaks at m/z 452.47, 508.1245, 580.4786, 608.6136, and 638.6136, respectively (Fig. S4–S9), supporting the proposed structures.
3.5 Electronic transitions and UV-vis spectral features
Magnetic moment measurements of metal ions are a valuable tool for determining electronic configurations and predicting molecular geometry.47 For the Co(II) complexes, the observed magnetic moment of 4.84 B.M. corresponds to three unpaired electrons, consistent with a spin-only distorted octahedral geometry. Magnetic susceptibility of the synthesized complexes was measured using the Gouy balance method, employing a calibrated Gouy tube and using Hg[Co(SCN)4] as the standard reference material. Measurements were carried out at room temperature, and the necessary diamagnetic corrections for both the ligand framework and metal ions were applied using Pascal's constants. The corrected molar susceptibility values were then used to calculate the effective magnetic moment (µ_eff) according to the standard equation µ_eff = 2.828(χ M T)1/2.The electronic spectra of the amide-containing macrocyclic ligands (N4O4MacL1–N4O4MacL3) display two prominent absorption bands in the ranges of 312–317 nm and 337–355 nm. The first band is attributed to the π → π* transitions within the amide chromophores,45 while the second band around 350 nm arises from n → π* transitions, superimposed on a charge-transfer process from the aromatic ring system to the amide group. Upon coordination with the metal center via the amide nitrogen atoms, coinciding with an electronic charge-transfer process originating from the aromatic ring and directed toward the amide functional group.
The electronic spectra and magnetic moment data of all Co(II) complexes recorded at room temperature are consistent with the proposed distorted octahedral geometry (Table 4). Specifically, the Co(II) complexes exhibit three d–d transition bands at 759–789 nm, 569–589 nm, and 439–461 nm, corresponding to the 4T1g → 4T2g (F), 4T1g → 4A2g, and 4T1g → 4T2g (P) transitions, respectively.46
| Compound | M (ohm−1 cm2 mol−1 | λmax (nm) |
|---|---|---|
| [N4O4MacL1] | — | 312, 359 |
| [N4O4MacL2] | — | 316, 341 |
| [N4O4MacL3] | — | 319, 349 |
| [Co(N4O4MacL1)Cl2] | 14.8 | 449, 589, 765 |
| [Co(N4O4MacL2)Cl2] | 15.9 | 439, 590, 795 |
| [Co(N4O4MacL3)Cl2] | 13.7 | 461, 580, 782 |
3.6 EPR
EPR spectroscopy was employed to investigate the presence of the paramagnetic electrons associated with the Co(II) coordination centers. The spectra of [Co(N4O4MacL1)Cl2]–[Co(N4O4MacL3)Cl2] were recorded in the solid state at liquid nitrogen temperature (77 K) and as polycrystalline samples dissolved in acetonitrile. Comparable g-values in solid-state and solution measurements confirm the presence of one unpaired electron in the dx2−y2 orbital. This observation aligns with a distorted octahedral stereochemistry for the Co(II) complexes.48The EPR data (Table S2) exhibited the characteristic relation g∥ > g⊥ > 2, consistent with axial symmetry. From these spectra, both g-values and the spin–orbit coupling parameter G were calculated. The shift relative to the free-electron g-value (2.0023) reflects appreciable orbital angular momentum involvement, corroborating the distorted octahedral geometry proposed for the Co(II) complexes.
3.7 Thermogravimetric analysis
Thermal analysis is commonly used to evaluate the thermal stability of ligands (N4O4MacL1–N4O4MacL3) and their Co(II) complexes ([Co(N4O4MacL1)Cl2]–[Co(N4O4MacL3)Cl2]) and to detect the presence of solvent molecules within the coordination sphere. Thermogravimetric analysis (TGA) was carried out for all synthesized compounds over the temperature range of 50–700 °C. The thermograms of the complexes exhibited similar patterns.Representative TGA curves of N4O4MacL1 and [Co(N4O4MacL1)Cl2] indicated no significant weight loss up to 160 °C, confirmed that no water molecules were present either inside or outside the coordination environment. The decomposition of [Co(N4O4MacL1)Cl2] occurred in three distinct stages. The first stage, between 230–430 °C, involved a mass loss of 19.39% (calcd. 20.08%), corresponding to the loss of two chloride ions. The second stage, from 160–210 °C, accounted for the decomposition of the organic ligand framework, resulting in a 44.35% mass loss (calcd. 46.78%).49 The third stage, observed between 570–640 °C, involved a further weight loss of 15.46% (calcd. 17.36%) due to the breakdown of the remaining coordinated ligand moiety. Above 640 °C, the TGA curve became nearly horizontal, indicating the formation of the final metal oxide (Fig. 4).
The experimentally observed mass losses for [Co(N4O4MacL1)Cl2] closely matched the calculated values based on its proposed chemical formula. These results demonstrate that the complexes are thermally stable up to 640 °C, and similar thermal behavior was observed for all other complexes.
 | (1) |
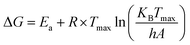 | (2) |
| ΔH = Ea + RTmax | (3) |
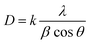 | (4) |
h = Planck constant (6.626 × 10−34 J s−1) and KB = Boltzmann constant (1.38 10−23 J s−1).
The data are presented in Tables 5 and 6. The results indicate that the values obtained from the two analytical methods are in close agreement (Fig. 5 and 6, S8–S11). The relatively low activation energy values for the Co(II) complexes suggest an autocatalytic effect of the metal ions on the thermal decomposition process. The observed negative entropy of activation (ΔS‡) values indicate a slow reaction, with the activated complex being more ordered than the initial reactants. This increased order in the transition state may be attributed to bond polarization, potentially facilitated by charge transfer electronic transitions during the decomposition process.
| Compounds | R | (Tmax) | (Ea) | A | ΔG | ΔH | ΔS |
|---|---|---|---|---|---|---|---|
| [N4O4MacL1] | 0.85383 | 571.18 | 13.532 | 0.00145 | 174.788 ± 0.65 | −4.115 ± 0.03 | −294.25 ± 1.8 |
| [N4O4MacL2] | 0.82104 | 760.39 | 16.657 | 0.00129 | 220.897 ± 0.72 | −6.225 ± 0.05 | −281.25 ± 2.1 |
| [N4O4MacL3] | 0.93138 | 540.49 | 36.654 | 0.00445 | 158.044 ± 0.58 | −4.248 ± 0.03 | −285.19 ± 1.7 |
| [Co(N4O4MacL1)Cl2] | 0.94602 | 734.28 | 15.673 | 0.00029 | 238.025 ± 0.74 | −6.654 ± 0.04 | −313.45 ± 2.3 |
| [Co(N4O4MacL2)Cl2] | 0.91324 | 612.18 | 180.12 | 0.00689 | 172.843 ± 0.63 | −4.514 ± 0.05 | −274.58 ± 1.9 |
| [Co(N4O4MacL3)Cl2] | 0.94214 | 541.13 | 356.58 | 0.0399 | 167.435 ± 0.57 | −4.320 ± 0.03 | −301.45 ± 2.0 |
| Compound | R | Tmax | Ea | A | ΔG | ΔH | ΔS |
|---|---|---|---|---|---|---|---|
| [N4O4MacL1] | 0.91245 | 574.33 | 18.465 | 0.0024 | 175.252 | −4.786 | −291.72 |
| [N4O4MacL2] | 0.9487 | 761.32 | 257.24 | 0.0155 | 218.148 | −6.205 | −279.73 |
| [N4O4MacL3] | 0.85845 | 544.42 | 21.520 | 0.0023 | 162.684 | −4.678 | −291.43 |
| [Co(N4O4MacL1)Cl2] | 0.93745 | 727.73 | 27.450 | 0.0019 | 225.402 | −6.754 | −294.82 |
| [Co(N4O4MacL2)Cl2] | 0.91248 | 612.92 | 46.536 | 0.0042 | 189.121 | −5.221 | −288.08 |
| [Co(N4O4MacL3)Cl2] | 0.95471 | 537.09 | 250.25 | 0.0344 | 144.765 | −4.188 | −265.74 |
 | ||
| Fig. 5 Coats–Redfern kinetic plots for macrocyclic ligand N4O4MacL1 and the corresponding Co(II) complex [Co(N4O4MacL1)Cl2]. | ||
3.8 PXRD analysis
The powder X-ray diffraction (XRD) patterns of the ligands (N4O4MacL1–N4O4MacL3) and their Co(II) complexes ([Co(N4O4MacL1)Cl2]–[Co(N4O4MacL3)Cl2]) are shown in Fig. 8. Multiple diffraction peaks indicate the polycrystalline nature of the samples. Crystals suitable for single-crystal XRD analysis could not be obtained for the Co(II) complexes. Therefore, the powder XRD data were used to investigate the crystal structures. The ligand [N4O4MacL1] (Fig. 7a) exhibited sharp, well-defined peaks, confirming its crystalline nature, whereas the complex [Co(N4O4MacL1)Cl2] (Fig. 8b) showed broader and less intense peaks, indicative of partial amorphous character. A close correlation was observed between the theoretical and experimental d-spacing values for the complexes. Crystallite dimensions were determined using the Debye–Scherrer formula (eqn (1)), employing the diffraction line of maximum intensity for the calculation.54 | ||
| Fig. 7 Powder X-ray diffraction profiles of (a) macrocyclic ligand N4O4MacL1 and (b) the corresponding cobalt(II) complex [Co(N4O4MacL1)Cl2]. | ||
 | ||
| Fig. 8 DFT-optimized molecular geometries of the macrocyclic ligand N4O4MacL1 and the corresponding Co(II) complex [Co(N4O4MacL1)Cl2]. | ||
The crystallite size (D) was calculated using the Debye–Scherrer equation, where k is the shape factor (0.94 for Cu Kα radiation), λ is the X-ray wavelength (1.5406 Å), θ is the Bragg diffraction angle, and β is the full width at half maximum (FWHM) of the diffraction peak. Using this approach, the crystallite size of the complex [Co(N4O4MacL1)Cl2] was determined to be 44.34 nm. Structural analysis indicates that [Co(N4O4MacL1)Cl2] adopts an orthorhombic crystal system. According to the result of powder X-ray diffraction studies, it has been found that the crystal system of macrocyclic complex [Co(N4O4MacL1)Cl2] orthorhombic with the unit cell parameters of a = 7.758, b = 17.542, c = 5.652 and α = β = γ = 90°, respectively. Table 7 reports the 2θ angles and Miller indices h, k, and l for each d value.
| Peak no. | 2θ (obs.) | 2θ (calc.) | h | k | l | d-spacing (obs.) | d-spacing (calc.) |
|---|---|---|---|---|---|---|---|
| 1 | 10.241 | 10.238 | 0 | 2 | 0 | 8.847 | 8.838 |
| 2 | 11.845 | 11.798 | 1 | 1 | 0 | 7.265 | 7.254 |
| 3 | 14.312 | 14.307 | 1 | 2 | 0 | 6.012 | 6.025 |
| 4 | 15.412 | 15.405 | 0 | 1 | 1 | 5.545 | 5.565 |
| 5 | 18.549 | 18.534 | 1 | 3 | 0 | 4.354 | 4.348 |
| 6 | 20.542 | 20.525 | 1 | 1 | 1 | 4.615 | 4.605 |
| 7 | 21.845 | 21.868 | 1 | 2 | 1 | 4.021 | 4.034 |
| 8 | 22.372 | 22.358 | 0 | 3 | 1 | 4.451 | 4.449 |
| 9 | 23.145 | 23.130 | 2 | 0 | 0 | 3.393 | 3.387 |
| 10 | 23.469 | 23.472 | 2 | 1 | 0 | 3.215 | 3.224 |
| 11 | 23.278 | 23.225 | 1 | 4 | 0 | 3.054 | 3.016 |
| 12 | 24.255 | 24.241 | 1 | 3 | 1 | 3.015 | 3.025 |
| 13 | 24.742 | 24.725 | 2 | 2 | 0 | 3.152 | 3.139 |
| 14 | 26.263 | 26.235 | 2 | 3 | 0 | 3.012 | 3.042 |
| 15 | 27.742 | 27.768 | 2 | 0 | 1 | 3.651 | 3.645 |
| 16 | 28.125 | 28.104 | 2 | 1 | 1 | 3.178 | 3.164 |
| 17 | 28.654 | 28.635 | 1 | 4 | 1 | 3.214 | 3.205 |
| 18 | 30.158 | 30.145 | 2 | 2 | 1 | 3.135 | 3.115 |
3.9 Molecular modelling
The molecular structures of the ligands (N4O4MacL1–N4O4MacL3) and their Co(II) complexes were modelled using the ORCA program in combination with Avogadro 4.0 software.55 Molecular mechanics methods were first applied to pre-optimize the regions of the molecules not directly coordinated to the metal centre. Energy minimization was performed iteratively for each molecule until convergence was achieved. In the optimized structures, the ligands adopt a highly folded conformation (Fig. 8), with the four oxygen atoms alternately displaced from the coordination plane constituted by the nitrogen atoms.For the [Co(N4O4MacL1)Cl2] complex, the four nitrogen donor atoms occupy the equatorial plane, creating a distorted octahedral geometry around the Co(II) centre (Fig. 9). The Co(II) ion is situated near the mean plane of the equatorial donor set. Comparison of bond lengths (Å) and bond angles (°) for the ligands and complexes (Tables 8 and 9) reveals variations that reflect the coordination interactions between the ligand donor atoms and the metal ion.53,54 The equatorial Co–N distances are 2.09, 2.29, and 2.05 Å, while the two axial Co–Cl bonds are measured at 2.298–2.299 Å. These values are consistent with those reported in the literature.56
 | ||
| Fig. 9 3D plots illustrating the HOMO and LUMO energy distributions for ligands N4O4MacL1–N4O4MacL3. | ||
| Compounds | (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
(C–N) | (N–H) | (Co–N) | (Co–Cl) |
|---|---|---|---|---|---|
| [N4O4MacL1] | 1.21232 | 1.35514 | 1.02512 | — | — |
| [N4O4MacL2] | 1.21295 | 1.36214 | 1.02114 | — | — |
| [N4O4MacL3] | 1.22104 | 1.36254 | 1.02385 | — | — |
| [Co(N4O4MacL1)Cl2] | 1.22363 | 1.44125 | 1.02745 | 2.11325 | 2.28979 |
| [Co(N4O4MacL2)Cl2] | 1.22145 | 1.45412 | 1.03954 | 2.35214 | 2.25347 |
| [Co(N4O4MacL3)Cl2] | 1.24125 | 1.43895 | 1.03745 | 2.06578 | 2.27412 |
| Atom-to-atom linkage | (Bond angles (°) | (Atomic connectivity) | (Bond angles (°) |
|---|---|---|---|
| N–Co–N | 89.2854 | N–Co–Cl | 88.9874 |
| N–Co–N | 88.1845 | N–Co–Cl | 90.4512 |
| N–Co–N | 178.1245 | N–Co–Cl | 91.0215 |
| N–Co–Cl | 86.4578 | N–Co–Cl | 83.1245 |
| N–Co–Cl | 93.5641 | Cl–Co–Cl | 176.4512 |
| N–Co–N | 178.3254 | ||
| N–Co–N | 91.1245 | ||
| N–Co–Cl | 88.5741 | ||
| N–Co–Cl | 91.0215 | ||
| N–Co–N | 91.0145 |
Frontier molecular orbitals, specifically the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO),60 play a key role in determining chemical stability. The HOMO represents the electron-donating ability of the molecule, while the LUMO corresponds to its electron-accepting capability. In addition, several global reactivity descriptors were calculated, including the frontier molecular orbital energy gap (ΔE) between HOMO and LUMO levels, the global descriptors χ, µ, η, S, and ω representing electronegativity, chemical potential, hardness, softness, and electrophilicity, respectively, and the maximum additional electronic charge (ΔN_max), with values reported in Table 10.
| Parameters | (N4O4MacL1) | (N4O4MacL2) | (N4O4MacL3) | 9(a) | 9(b) | 9(c) |
|---|---|---|---|---|---|---|
| EHOMO (eV) | −7.358 | −7.547 | −7.145 | −6.754 | −6.124 | 6.245 |
| ELUMO (eV) | −1.541 | −1.812 | −1.325 | −2.214 | −2.147 | −2.149 |
| ΔE (eV) | 6.124 | 5.504 | 5.0154 | 4.387 | 3.0215 | 4.213 |
| IE (eV) | 7.541 | 7.648 | 7.745 | 6.145 | 6.358 | 6.345 |
| χ (eV) | 4.314 | 4.4123 | 4.2154 | 4.845 | 4.6587 | 4.4045 |
| η (eV) | 3.145 | 2.8745 | 2.7945 | 2.124 | 1.7458 | 2.1254 |
| S (eV)−1 | 0.1287 | 0.1145 | 0.0158 | 0.2154 | 0.2584 | 0.2545 |
| ω (eV) | 3.5412 | 3.7452 | 4.048 | 5.5845 | 5.1248 | 4.2541 |
| Pi | −4.625 | −4.2514 | −4.6145 | −4.336 | −4.5142 | −4.1478 |
| Nmax | 1.4875 | 1.2014 | 1.2548 | 2.0987 | 2.0215 | 2.32514 |
The HOMO–LUMO energy gap (ΔE = 4.49 eV) is an important parameter reflecting molecular stability; higher values indicate greater stability. The negative chemical potential (µ) combined with a positive electrophilicity index (ω) suggests that the amide-based ligands are capable of donating electrons to metal centers, supporting their role in complex formation.
3.10 Biological activities
| Compounds | Inhibition zone (mm) | |||||||
|---|---|---|---|---|---|---|---|---|
| B. subtilis | S. aureus | E. coli | S. Typhi | |||||
| Conc. (µg ml−1) | 30.0 | 50 | 30.0 | 50.0 | 30.0 | 50.0 | 30.0 | 50.0 |
| [N4O4MacL1] | 8 | 9 | 6 | 8 | 10 | 11 | 7 | 9 |
| [N4O4MacL2] | 10 | 11 | 8 | 9 | 12 | 13 | 10 | 10 |
| [N4O4MacL3] | 9 | 10 | 7 | 8 | 13 | 17 | 9 | 11 |
| [Co(N4O4MacL1)Cl2] | 10 | 11 | 9 | 10 | 19 | 20 | 13 | 14 |
| [Co(N4O4MacL2)Cl2] | 12 | 14 | 10 | 11 | 18 | 21 | 15 | 16 |
| [Co(N4O4MacL3)Cl2] | 14 | 17 | 9 | 10 | 17 | 19 | 14 | 17 |
| Ampicillin | 16 | 20 | 11 | 13 | 24 | 26 | 18 | 20 |
The enhanced antibacterial activity of the Co(II) complexes can be rationalized using Tweedy's chelation theory and Overtone's concept of cell permeability. Chelation reduces the polarity of the central metal ion due to partial sharing of its positive charge with the ligand donor atoms, which in turn increases the lipophilicity of the complex. Increased lipophilicity facilitates the penetration of the complex through the lipid bilayer of bacterial cell membranes, consistent with Overtone's idea that only lipid-soluble molecules efficiently cross cell membranes. Enhanced electron delocalization within the chelate ring further supports membrane permeability. Once inside the cell, the metal complexes can interact with bacterial enzymes, potentially inhibiting key biochemical pathways such as respiration, ultimately leading to cell death.63–65 The relative antibacterial potency of the complexes follows the order: [Co(N4O4MacL2)Cl2] > [Co(N4O4MacL3)Cl2] > [Co(N4O4MacL1)Cl2] (Fig. 11).
The antifungal activity of the compounds was evaluated in vitro against Aspergillus niger and Candida albicans using the serial dilution method. The minimum inhibitory concentration (MIC) values are listed in Table 12. Comparison of the MIC data indicates that the Co(II) complexes display greater antifungal activity than the free ligands. Among the tested compounds, [Co(N4O4MacL2)Cl2] exhibited the highest antifungal efficacy relative to the other complexes and the standard antifungal agent (Fig. 12).
| Compounds | Fungal inhibition % (conc. in µg ml−1) | |||
|---|---|---|---|---|
| A. Niger | C. albicans | |||
| Conc. (µg ml−1) | 500 (µg ml−1) | 1000 (µg ml−1) | 500 (µg ml−1) | 1000 (µg ml−1) |
| [N4O4MacL1] | 28 | 39 | 23 | 31 |
| [N4O4MacL2] | 31 | 43 | 24 | 32 |
| [N4O4MacL3] | 19 | 41 | 22 | 32 |
| [Co(N4O4MacL1)Cl2] | 58 | 71 | 39 | 55 |
| [Co(N4O4MacL2)Cl2] | 61 | 75 | 43 | 57 |
| [Co(N4O4MacL3)Cl2] | 60 | 74 | 41 | 66 |
| Nystatin | 69 | 84 | 53 | 68 |
 | ||
| Fig. 15 Comparative cytotoxic evaluation of macrocyclic ligands (N4O4MacL1–N4O4MacL3) and the corresponding Co(II) complexes. | ||
| Compounds | MCF-7 | HepG-2 |
|---|---|---|
| [N4O4MacL1] | 65.01 ± 0.19 | 50.42 ± 0.66 |
| [N4O4MacL2] | 61.20 ± 0.18 | 47.23 ± 0.57 |
| [N4O4MacL3] | 63.11 ± 0.21 | 49.41 ± 0.61 |
| [Co(N4O4MacL1)Cl2] | 20.13 ± 0.31 | 21.38 ± 0.13 |
| [Co(N4O4MacL2)Cl2] | 18.42 ± 0.25 | 17.94 ± 0.45 |
| [Co(N4O4MacL3)Cl2] | 21.71 ± 0.45 | 18.62 ± 0.49 |
| Colchicine | 16.80 ± 0.02 | 10.00 ± 0.02 |
Importantly, the observed cytotoxic activity correlates well with predictions from molecular docking studies.69,70 The colorimetric viability assays further indicate that both the ligands and their Co(II) complexes exhibit cytotoxic effects against MCF-7 cells comparable to colchicine (Fig. 15), with Co(II) complexes showing higher average levels of relative cell inhibition. Fig. 16 depicts the survival curves of the cancer cell lines treated with the synthesized compounds.
3.11 Computational molecular docking analysis
To investigate the protein-binding potential of the synthesized ligands computationally, molecular docking studies were performed to evaluate their interaction with various receptor proteins. These studies provided binding affinities and inhibitory constants, indicating that most ligands exhibited significant binding potential. Docking simulations were carried out using the AutoDock Vina module within PyRx software.71 Protein structures in PDB format (3T88, 3TY7, 3DRA, 5H67) were loaded as N4O4Macromolecules, while ligand preparation involved generating different tautomer's, assigning bond orders, defining ring conformations, and specifying stereochemistry. These targets were chosen based on their relevance to the pathogenic organisms tested in our antimicrobial studies and their biological role in essential cellular pathways, such as cell wall synthesis, enzyme regulation, nucleic acid processing, and oxidative stress response. Each protein represents a clinically significant therapeutic target associated with resistance development or virulence. Furthermore, previously reported studies have demonstrated the involvement of these proteins in pathogen survival, making them suitable receptors for molecular docking to evaluate potential inhibitory interactions of the synthesized macrocycles and metal complexes (Fig. 17).72 | ||
| Fig. 17 (a) Two-dimensional and three-dimensional interaction profiles of N4O4MacL1 with the E. coli protein receptor. | ||
For Escherichia coli, the crystal structure 3T88 (ref. 73) was employed for docking, which is critical for assessing protein function related to cell growth and viability. The best-docked conformation of ligand N4O4MacL1 exhibited a binding score of −12.8 kcal mol−1 (Table 14), indicating strong inhibition. The ligand formed two conventional hydrogen bonds with ARG169 and ARG1293, with bond lengths of 2.41 Å and 2.42 Å, respectively (Fig. 18). Docking of N4O4MacL1–N4O4MacL3 ligands yielded minimal binding energies of −12.8, −10.3, and −11.5 kcal mol−1, respectively, highlighting N4O4MacL1 as the ligand with the strongest predicted interaction.
| Compounds | Active protein (PDB ID) | Binding affinity (kcal mol−1) | Number of H-bond | Binding mode |
|---|---|---|---|---|
| [N4O4MacL1] | 3T88 | −12.8 | 2 | ARG′169, AGG′1293 |
| 3ROW | −9.9 | 1 | ALA′497 | |
| 3DRA | −9.8 | 1 | TYR′550 | |
| 5H67 | −9.6 | 3 | ARG′54, ARG′45, SER′142 | |
| [N4O4MacL2] | 3T88 | −10.3 | 1 | ARG′60 |
| 3ROW | −9.7 | 4 | ASP′487, GLU′485,GLY428, HIE′457 | |
| 3DRA | −11.3 | 2 | ARG′615, HIE′425 | |
| 5H67 | −10.8 | 3 | GLU′50, LYS′135, SER′52 | |
| [N4O4MacL3] | 3T88 | −11.5 | 1 | ARG′341 |
| 3ROW | −10.3 | 3 | GLU′103, ASN′627, THR′859 | |
| 3DRA | −11.2 | 2 | MET′624, PRO′623 | |
| 5H67 | −9.7 | 1 | GLU′50 |
 | ||
| Fig. 18 (a) Two-dimensional and three-dimensional interaction profiles of N4O4MacL1 with the 3row protein receptor. | ||
Docking with the N4O4Macromolecule 3ROW revealed hydrogen bonding interactions of the ligands with GLU485, ASP487, HIE457, and GLY428,74 at bond distances of 2.50, 2.59, 3.01, and 2.89 Å, respectively (Fig. 18). The corresponding binding energies for N4O4MacL1–N4O4MacL3 were −9.8, −9.8, and −10.4 kcal mol−1, with N4O4MacL3 showing the strongest negative binding energy, indicating a more favorable interaction with the protein. Similar docking analyses were performed for PDB structures 3DRA and 5H67, as depicted in Fig. 19 and 20. Overall, these docking results correlate well with experimental observations from spectroscopic studies, supporting the proposed groove-binding mode of the ligands to the target biomolecules.
 | ||
| Fig. 19 (a) Two-dimensional and three-dimensional interaction profiles of N4O4MacL2 with the 3dra protein receptor. | ||
 | ||
| Fig. 20 (a) Two-dimensional and three-dimensional interaction profiles of N4O4MacL1 with the 5h67 protein receptor. | ||
4 Conclusion
The Co(II) complex derived from 1,8-diaminonaphthalene was successfully synthesized and characterized using a combination of analytical and multi-spectroscopic techniques, confirming its biochemical and physiological activity. FT-IR analysis indicated coordination through the amide nitrogen atoms, validating ligand donation to the central metal. The proposed distorted octahedral geometry of the Co(II) complex was aadditionally validated through complementary UV-visible, ESI-MS, and EPR spectroscopic evidence. DFT calculations corroborated the predicted structures of the synthesized compounds. In vitro cytotoxicity studies demonstrated that [Co(N4O4MacL2)Cl2] and [Co(N4O4MacL1)Cl2] showed pronounced anticancer efficacy against MCF-7 cells, yielding IC50 values of 19.35 ± 0.23 µM and 21.54 ± 0.30 µM, respectively. In addition, the synthesized compounds demonstrated significant antimicrobial potential, showing effective inhibition against both bacterial and fungal pathogens. Molecular docking studies revealed favorable binding energies, suggesting potential biological targets for the complexes. Specifically, [Co(N4O4MacL3)Cl2] exhibited strong antimicrobial activity, showing a minimum inhibitory concentration of 67 µg mL−1 toward Candida albicans. In addition, antioxidant studies demonstrated excellent radical-quenching efficiency for [Co(N4O4MacL2)Cl2], reflected by an IC50 value of 35.08 ± 1.22 µg mL−1. Docking simulations further indicated that the Macrocyclic ligand N4O4MacL3 displayed higher negative binding affinities toward receptors 3T88, 3DRA, 3ROW, and 5H67, supporting the observed biological activities. These results collectively suggest that the amide-based ligands and their Co(II) complexes are promising candidates for antimicrobial, anticancer, and antioxidant applications.Author contributions
The research concept was proposed by Subhash and Ashu Chaudhary. Experimental work was carried out by Manish Kumar and Vandana. Subhash performed data analysis, manuscript preparation, and editing. All authors critically reviewed and approved the final manuscript.Conflicts of interest
The authors declare no competing interests.Data availability
All data are available in the article and its supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra08873a.Acknowledgements
The author (Subhash) expresses their heartfelt gratitude to Smt. Devkiba Mohansinhji Chauhan College of Commerce and Science, Silvassa, for extending laboratory facilities, research infrastructure, and continuous academic encouragement throughout the duration of this study. The authors sincerely thank the Indian Institute of Science (IISc), Bengaluru, India, for providing the cancer cell line necessary for carrying out this research work. The support and motivation provided by the college administration and faculty members are gratefully appreciated. The authors express sincere appreciation to the DST-FIST Program 2017 (Proposal No. SR/FST/CS-I/2017/12(C), sanctioned on 10.05.2018) for providing financial and infrastructural support at Kurukshetra University, Kurukshetra.References
- J. Ferlay, H. R. Shin, F. Bray, D. Forman, C. Mathers and D. M. Parkin, Int. J. Cancer, 2010, 127, 2893 Search PubMed
.
- S. Kumar, J. Devi, A. Dubey, D. Kumar, D. K. Jindal, S. Asija and A. Sharma, Res. Chem. Intermed., 2023, 49(3), 939 CrossRef CAS
.
- A. Erdmann, Y. Menon, C. Gros, V. Masson, Y. Aussagues, F. Ausseil, N. Novosad, P. Schambel, M. Baltas and P. B. Arimondo, Future Med. Chem., 2016, 8(4), 373 CrossRef CAS
.
- W. Khalid, A. Badshah, A. UllahKhan, H. Nadeem and S. Ahmed, Chem. Cent. J., 2018, 12, 1 CrossRef PubMed
.
- M. Gaber, A. M. Khedr and M. Elsharkawy, Appl. Organomet. Chem., 2017, 31(12), e3884 CrossRef
.
- A. Alka, S. Gautam, R. Kumar, P. Singh, N. Gandhi and P. Jain, Results Chem., 2023, 5, e100849 CrossRef
.
- V. R. Chandrasekhar, K. M. Palsamy, R. Lokesh, D. Thangadurai, I. N. Gandhi, R. Jegathalaprathaban and R. Gurusamy, Appl Organometal Chem., 2019, 33, e4753 CrossRef
.
- H. Kargara, R. Behjatmanesh-Ardakanib, V. Torabib, A. Sarvianb, Z. Kazemic, Z. Chavoshpour-Natanzic, V. Mirkhanic, A. Sahraeid, M. N. Tahire and M. Ashfaq, Inorg. Chim. Acta, 2021, 514, 120004 Search PubMed
.
- X. Liu and J. R. Hamon, Coord. Chem. Rev., 2019, 389, 94 CrossRef CAS
.
- N. Sharma, N. Dhingra, G. Choudhary, S. Chauhan and H. L. Singh, Integrated Spectroscopic, Computational, and Antimicrobial Profiling of Phenylsilicon (IV) Schiff Base Complexes, J. Indian Chem. Soc., 2025, 102216 CrossRef CAS
.
- D. Ghosh, S. T. Choudhury, S. Ghosh, A. K. Mandal, S. Sarkar, A. Ghosh, K. D. Saha and N. Das, Chem Biol Interact, 2012, 195(3), 206 CrossRef CAS PubMed
.
- M. Jafari, M. Salehi, M. Kubicki, A. Arab and A. Khaleghian, Inorg. Chim. Acta, 2017, 462, 329 Search PubMed
.
- A. A. Mohamed, F. M. Ahmed, W. A. Zordok, W. H. El-Shwiniy, S. A. Sadeek and H. S. Elshafe, Inorganics, 2022, 10(11), 177 Search PubMed
.
- M. S. El-Attar, F. M. Ahmed, S. A. Sadeek, S. F. Mohamed, W. A. Zordok and W. H. El-Shwiniy, Appl. Organomet. Chem., 2022, 36(10), 36826 CrossRef
.
- N. Sharma, N. Dhingra and H. L. Singh, Design, spectral, antibacterial and in-silico studies of new thiosemicarbazones and semicarbazones derived from symmetrical chalcones, J. Mol. Struct., 2024, 1307, 138000 CrossRef CAS
.
- W. A. Wani, A. H. Malik, A. Hussain, M. F. Alajmi, S. Shreaz and I. Kostova, Recent advances in metal complexes for the photodynamic therapy of cancer, New J. Chem., 2025, 49(31), 13244–13268 RSC
.
- H. A. El-Boraey and S. A. Aly, Syn. React, Inorg. Met., 2013, 43, 1130–1138 CAS
.
- M. Cocu, I. Bulhac, E. Coropceanu, E. Melnic, S. Shova, O. Ciobanica, V. Gutium and P. Bourosh, J. Mol. Struct., 2014, 1063, 274–282 CrossRef CAS
.
- P. Rawat, R. N. Singh, A. Ranjan, S. Ahmad and R. Saxena, Spectrochim. Acta, 2017, 179, 1–10 CrossRef CAS PubMed
.
- S. Karadeniz, C. Yuksektepe Ataol, T. Ozen, R. Demir, H. Ogütçü and H. Bati, J. Mol. Struct., 2019, 1175, 39–48 CrossRef CAS
.
- R. J. Wani, A. Hussain, M. U. D. Sheikh, M. N. Bukhari, M. Fatima and G. Ali, G. Synthesis of mono and hetero-bi nuclear lanthanum (III) and europium (III) complexes of 2-hydroxy-5-sulfobenzoic acid and 1, 10-phenanthroline: luminescence and magnetic studies, J. Mater. Sci. Mater. Eng., 2025, 20(1), 64 CrossRef
.
- B. Jeragh, M. S. Ali and A. A. El-Asmy, Spectrochim. Acta, 2015, 145, 295–301 CrossRef CAS PubMed
.
- Y. Burgos-Lopez, J. Del Pla, L. M. Balsaa, I. E. Leon, G. A. Echeverri, O. E. Piro, J. Garcia-Tojal, R. Pis-Diez, A. C. Gonzalez-Baro and B. S. Parajon-Costa, Inorg. Chim. Acta, 2019, 487, 31–40 CrossRef CAS
.
- R. Bhaskar, N. Salunkhe, A. Yaul and A. Aswar, Spectrochim, Acta, 2015, 151, 621–627 CrossRef CAS PubMed
.
- J. Subhash and A. Chaudhary, Res. Chem. Intermed., 2023, 49, 4729–4758 CrossRef
.
- A. Subhash, J. Chaudhary, M. Kumar and R. Solanki, J. Iran. Chem. Soc., 2023, 20, 2339–2362 CrossRef
.
- M. Khodiev, U. Holikulov, A. Jumabaev, N. Issaoui, L. N. Lvovich, O. M. Al-Dossary and L. G. Bousiakoug, J. Mol. Liq., 2023, 382, e121960 CrossRef
.
- W. H. Mahmoud, R. G. Deghadi and G. G. Mohamed, J. Therm. Anal. Calorim., 2016, 127(3), 2149–2171 CrossRef
.
- H. A. Rudbari, M. R. Iravani, V. Moazam, B. Askari, M. Khorshidifard, N. Habibi and G. Bruno, J. Mol. Struct., 2016, 1125, 113–120 CrossRef
.
- H. L. Singh, N. Dhingra and S. Bhanuka, Synthesis, spectral, antibacterial and QSAR studies of tin and silicon complexes with Schiff base of amino acids, J. Mol. Struct., 2023, 1287, 135670 CrossRef CAS
.
- W. A. Wani, M. F. Alajmi, A. Hussain, and I. A. Wani, Therapeutic Potential of Gold Complexes, Royal Society of Chemistry. 2025 Search PubMed
.
- D. S. Bose, M. Idrees, N. M. Jakka and J. V. Rao, J. Comb. Chem., 2010, 12, 100–110 CrossRef CAS
.
- A. M. Abu-Dief, L. H. Abdel-Rahman and A. A. H. Abdel-Mawgoud, Appl. Organomet. Chem., 2020, 34(2), e5373 CrossRef CAS
.
- N. El-Metwaly, H. Katouah, E. Aljuhani, A. Alharbi, F. Alkhatib, M. Aljohani, S. Alzahrani, M. Y. Alfaifi and A. M. Khedr, J. Inorg. Organomet. Polym. Mater., 2020, 30(10), 4142–4154 CrossRef CAS
.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785 CrossRef CAS
.
- S. Subhash, A. Chaudhary, M. Mamta and M. Jyoti, Chem. Pap., 2023, 77, 5059–5078 CrossRef
.
- G. A. Giffin, A. Moretti, S. Jeong and S. Passerini, J. Phys. Chem. C, 2014, 118(19), 9966–9973 CrossRef CAS
.
- M. A. I. Al-Gaber, H. M. Abd El-Lateef, M. M. Khalaf, S. Shaaban, M. Shawky, G. Mohamed, A. Abdou, M. Gouda and A. M. Abu-Dief, Materials, 2023, 16(3), 897 CrossRef CAS PubMed
.
- A. Abdou, H. M. Mostafa and A. M. M. Abdel-Mawgoud, Inorg. Chim. Acta, 2022, 539, 121043 CrossRef CAS
.
- M. Malhotra, R. K. Rawal, D. Malhotra, R. Dhingra, A. Deep and P. C. Sharma, Arab. J. Chem., 2017, 10, 1022–1031 CrossRef
.
- M. Mishra, K. Tiwari, S. Shukla, R. Mishra and V. P. Singh, Spectrochim. Acta A Mol. Biomol. Spectrosc., 2014, 132, 452–464 Search PubMed
.
- H. Purandara, S. Forob and B. T. Gowdaa, Acta. Crystallogr. E., 2015, 71(7), 795–798 CAS
.
- W. H. Mahmoud, G. G. Mohamed and M. M. I. El-Dessouky, Int. J. Electrochem. Sci., 2014, 9(3), 1415–1438 Search PubMed
.
- A. H. Manikshete, M. M. Awatade, S. K. Sarsamkar and M. R. Asabe, Int. J. Eng. Sci. Inv., 2015, 4(1), 22–29 Search PubMed
.
- D. X. West, Y. Yang, T. L. Klein, K. I. Goldberg, A. E. Liberta, J. Valdes Martinez and S. Hernandez-Ortega, Polyhedron, 1995, 14, 3051–3060 CrossRef CAS
.
- M. S. Fataftah, M. D. Krzyaniak, B. Vlaisavljevich, M. R. Wasielewski, J. M. Zadrozny and D. E. Freedman, Chem. Sci., 2019, 10, 6707–6714 RSC
.
- S. Chandra, A. Gautam and M. Tyagi, J. Trans Met chem., 2007, 32, 1079–1084 CrossRef CAS
.
- P. Kopel, Z. Travnicek, J. Marek, M. Korabik and J. Mrozinski, Polyhedron, 2003, 22, 411 CrossRef CAS
.
- N. Kavitha and P. V. J. Anantha Lakshmi, Saudi Chem. Soc., 2017, 21, 457–463 CrossRef
.
- A. W. Coats and J. P. Redfern, Nature, 1964, 20, 68–79 CrossRef
.
- H. W. Horowitz and G. Metzger, Anal. Chem., 1963, 35, 1464–1468 CrossRef CAS
.
- S. Jyothi, K. Sreedhar, D. Nagavaju and S. J. Swamy, Can Chem Trans., 2015, 3(4), 368–380 CAS
.
- M. Tyagi and S. Chandra, Open J. Inorg. Chem., 2012, 2(3), 41–48 CrossRef
.
- M. K. Koley, O. P. Chouhan, S. Biswas, J. Fernandes, A. Banerjee, A. Chattopadhyay, B. Varghese, P. T. Manoharan and A. P. Koley, Inorg. Chim. Acta, 2017, 456, 179–198 CrossRef CAS
.
- Y. A. Alghuwainem, H. M. Abd El-Lateef, M. M. Khalaf, A. A. Abdelhamid, A. Alfarsi, M. Gouda and A. Abdou, J. Mol. Liq., 2023, 369, 120936 CrossRef CAS
.
- M. Kumar, P. J. Darolia, S. Chauhan, M. Sindhu, K. K. Verma and S. Garg, ChemistrySelect, 2021, 6(23), 5778–5790 CrossRef CAS
.
- S. Subhash, A. Phor, A. Chaudhary and M. Jyoti, J. Inorg. Organomet. Polym., 2024, 50(11), 1081–1111 Search PubMed
.
- M. Kumar, P. J. Darolia, S. Chauhan, M. Sindhu, K. K. Verma and S. Garg, ChemistrySelect, 2021, 6(23), 5778–5790 CrossRef CAS
.
- W. H. Mahmoud, N. F. Mahmoud and G. G. Mohamed, Applied Organomet. Chem, 2017, 31, 1 Search PubMed
.
- W. H. Mahmoud, G. G. Mohamed and A. M. Refat, Applied Organomet., 2017, 31(11), e3753 Search PubMed
.
- N. Raman, J. D. Raja and A. Sakthivel, J. Chem. Sci., 2007, 119, 303–310 CrossRef CAS
.
- M. A. Latif, T. Ahmed, M. S. Hossain, B. M. Chaki, A. Abdou and M. Kudrat-E-Zahan, Russ. J. Gen. Chem., 2023, 93(2), 389–397 Search PubMed
.
- A. Subhash, J. Chaudhary, M. Kumar, N. Kumar and N. K. Agarwal, J. Chem. Sci., 2022, 134(4), 113 CrossRef
.
- N. Dharmaraj, P. Viswanathamurthi and K. Natarajan, Transition Met. Chem., 2001, 26, 105–109 CrossRef CAS
.
- M. Akki, D. S. Reddy, K. S. Katagi, A. Kumar, H. C. Devarajegowda, V. Babagond, S. Mane and S. D. Joshi, J. Mol. Struct., 2022, 1266, 133452 Search PubMed
.
- P. Gull, M. A. Malik, O. A. Dar and A. A. Hashmi, Microb. Pathog., 2017, 104, 212–216 CrossRef CAS PubMed
.
- P. Rathi and D. P. Singh, J. Mol. Struct., 2015, 1100, 208–214 CrossRef CAS
.
- J. Haribabu, G. R. Subhashree, S. Saranya, K. Gomathi, R. Karvembu and D. Gayathri, J. Mol. Struct., 2015, 1094, 281–291 CrossRef CAS
.
- S. Y. Ebrahimipour, I. Sheikhshoaie, J. Castro, W. Haase, M. Mohamadi, S. Foro, M. Sheikhshoaie and S. E. A. Mahani, Inorg. Chim. Acta, 2015, 430, 245–252 Search PubMed
.
- A. Sharma, N. Dhingra, H. L. Singh, S. Khaturia and U. Bhardawaj, New Complexes of organotin (IV) and organosilicon (IV) with 2-{(3, 4-dimethoxybenzylidene) amino}-benzenethiol: Synthesis, spectral, theoretical, antibacterial, docking studies, J. Mol. Struct., 2022, 1261, 132812 CrossRef CAS
.
- A. Subhash, J. Chaudhary, M. Kumar and R. Solanki, J. Iran. Chem. Soc., 2023, 20, 2339–2362 Search PubMed
.
- M. A. Arafath, F. Adam, M. B. K. Ahamed, M. R. Karim, M. N. Uddin, B. M. Yamin and A. Abdou, J. Mol. Struct., 2023, 1278, 134887 Search PubMed
.
- M. E. Elshakre, A. Mahmoud, H. Noamaan and H. Butt Moustafa, Int. J. Mol. Sci., 2020, 21, 1253 CrossRef CAS PubMed
.
- A. Kumar and U. Bora, Interdiscip. Sci. Comput. Life Sci., 2014, 6, 285–291 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |